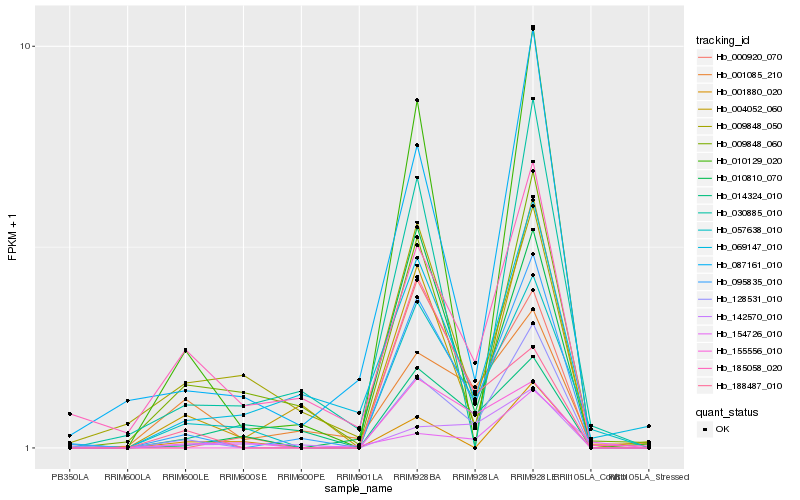
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_057638_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637008 [Jatropha curcas] |
| 2 |
Hb_000920_070 |
0.2091197983 |
- |
- |
- |
| 3 |
Hb_128531_010 |
0.2114768149 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_095835_010 |
0.2152747202 |
- |
- |
hypothetical protein JCGZ_23410 [Jatropha curcas] |
| 5 |
Hb_010810_070 |
0.2221117487 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_009848_060 |
0.2303102991 |
- |
- |
unknown [Glycine max] |
| 7 |
Hb_014324_010 |
0.2376107986 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 8 |
Hb_154726_010 |
0.2435610033 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 9 |
Hb_009848_050 |
0.2451768412 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK13 [Elaeis guineensis] |
| 10 |
Hb_001085_210 |
0.2562000608 |
- |
- |
PREDICTED: myosin-binding protein 3 [Jatropha curcas] |
| 11 |
Hb_004052_060 |
0.2573658597 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_069147_010 |
0.2587010659 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 13 |
Hb_087161_010 |
0.2608804796 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_185058_020 |
0.2608808718 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 15 |
Hb_155556_010 |
0.2630229571 |
- |
- |
putative methylenetetrahydrofolate reductase [Morus notabilis] |
| 16 |
Hb_188487_010 |
0.2667223636 |
- |
- |
hypothetical protein JCGZ_24382 [Jatropha curcas] |
| 17 |
Hb_142570_010 |
0.2692971338 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_001880_020 |
0.2696996473 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 19 |
Hb_010129_020 |
0.2720289299 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_030885_010 |
0.2754882689 |
- |
- |
- |