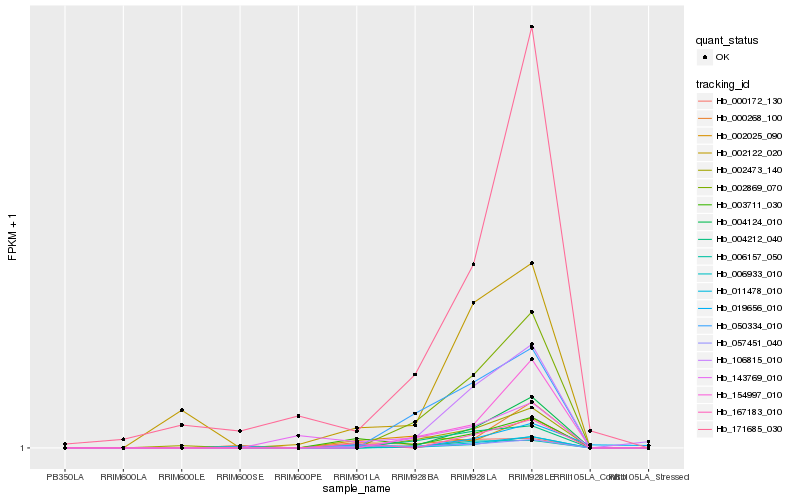
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011478_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003711_030 |
0.0309326102 |
- |
- |
PREDICTED: uncharacterized protein LOC102621950 [Citrus sinensis] |
| 3 |
Hb_167183_010 |
0.1766926093 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_057451_040 |
0.1831727786 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000268_100 |
0.2073534196 |
- |
- |
- |
| 6 |
Hb_002025_090 |
0.2274045082 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_002122_020 |
0.260048603 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 8 |
Hb_002869_070 |
0.2692897577 |
- |
- |
- |
| 9 |
Hb_006933_010 |
0.2758122577 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_004124_010 |
0.2762222906 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 11 |
Hb_004212_040 |
0.2764387991 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_143769_010 |
0.2829622059 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 13 |
Hb_019656_010 |
0.2896983953 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_154997_010 |
0.2903747129 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 15 |
Hb_106815_010 |
0.2916204766 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 16 |
Hb_050334_010 |
0.2986411249 |
- |
- |
- |
| 17 |
Hb_006157_050 |
0.2992136324 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000172_130 |
0.2994279736 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 19 |
Hb_171685_030 |
0.3004729684 |
- |
- |
URF-1 [Citrullus lanatus] |
| 20 |
Hb_002473_140 |
0.3011981441 |
- |
- |
- |