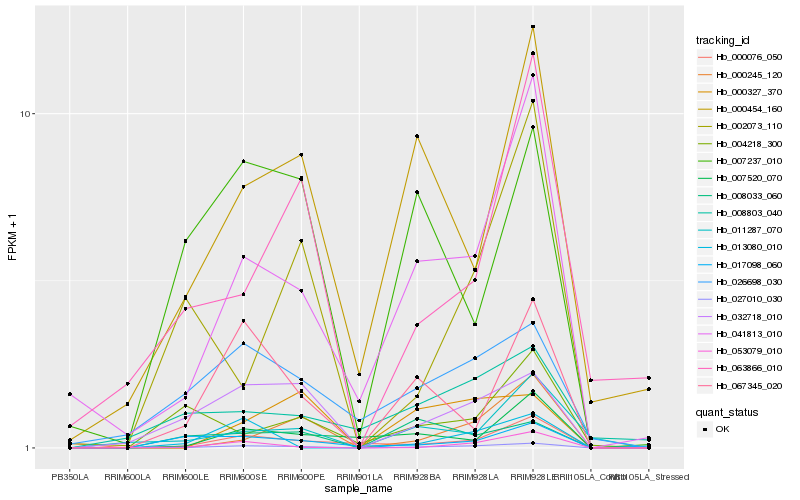
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027010_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_032718_010 |
0.1956116972 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_041813_010 |
0.2085801168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_026698_030 |
0.2399405956 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 5 |
Hb_053079_010 |
0.2492898301 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_067345_020 |
0.2516710571 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 7 |
Hb_017098_060 |
0.2640660185 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Pyrus x bretschneideri] |
| 8 |
Hb_000454_160 |
0.2657594487 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000245_120 |
0.2666733874 |
- |
- |
hypothetical protein POPTR_0001s18890g [Populus trichocarpa] |
| 10 |
Hb_000076_050 |
0.2714279317 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 11 |
Hb_007237_010 |
0.2759092593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011287_070 |
0.2764137313 |
- |
- |
- |
| 13 |
Hb_007520_070 |
0.2768445589 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 14 |
Hb_013080_010 |
0.2785930682 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_000327_370 |
0.2792067433 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 16 |
Hb_008033_060 |
0.2802660645 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 17 |
Hb_008803_040 |
0.2818021595 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |
| 18 |
Hb_004218_300 |
0.2840936176 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 19 |
Hb_002073_110 |
0.2860399298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_063866_010 |
0.2903697628 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |