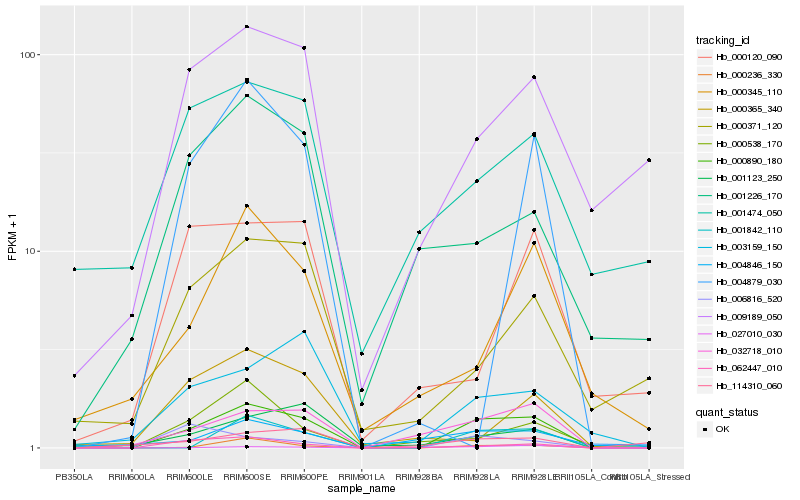
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_180 |
0.0 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 2 |
Hb_032718_010 |
0.2279844967 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_000236_330 |
0.2435365642 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001842_110 |
0.2497489055 |
transcription factor |
TF Family: ERF |
CBF-like transcription factor, putative [Ricinus communis] |
| 5 |
Hb_062447_010 |
0.2534294562 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 6 |
Hb_001123_250 |
0.2734564996 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |
| 7 |
Hb_003159_150 |
0.2763188954 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 8 [Jatropha curcas] |
| 8 |
Hb_009189_050 |
0.2898761886 |
- |
- |
PREDICTED: uncharacterized protein LOC105648080 [Jatropha curcas] |
| 9 |
Hb_000345_110 |
0.2943516065 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 10 |
Hb_114310_060 |
0.3004612928 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2G, putative [Ricinus communis] |
| 11 |
Hb_004846_150 |
0.3005755605 |
- |
- |
hypothetical protein JCGZ_26968 [Jatropha curcas] |
| 12 |
Hb_000371_120 |
0.3016292027 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000538_170 |
0.3068388554 |
- |
- |
PREDICTED: butyrate--CoA ligase AAE11, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_027010_030 |
0.3097915987 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_000365_340 |
0.314916306 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 16 |
Hb_001226_170 |
0.3165538553 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_006816_520 |
0.3172135255 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 18 |
Hb_000120_090 |
0.3192062097 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 19 |
Hb_004879_030 |
0.3207463334 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 20 |
Hb_001474_050 |
0.3222156174 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |