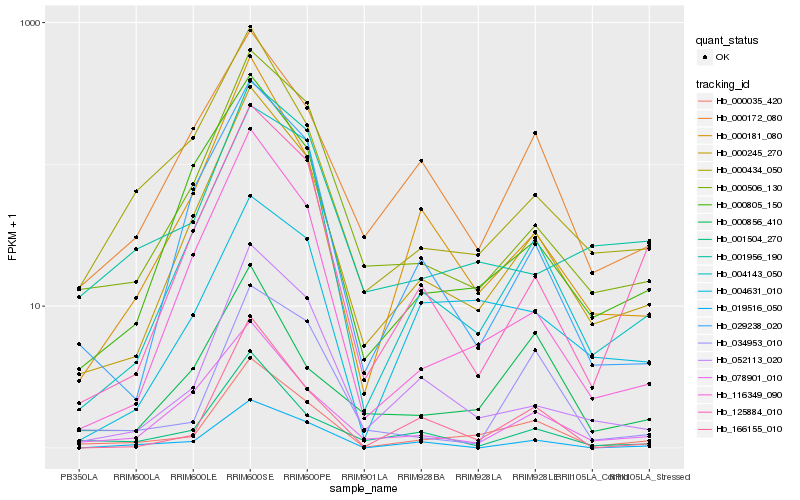
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_420 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 2 |
Hb_000245_270 |
0.1391470833 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 3 |
Hb_004143_050 |
0.1643905661 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 4 |
Hb_000506_130 |
0.1759837852 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 5 |
Hb_000172_080 |
0.1829355645 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 6 |
Hb_166155_010 |
0.1833167638 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 7 |
Hb_034953_010 |
0.186192358 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 8 |
Hb_004631_010 |
0.187221337 |
- |
- |
Early nodulin 20 precursor, putative [Ricinus communis] |
| 9 |
Hb_000181_080 |
0.1898557275 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 10 |
Hb_001504_270 |
0.1903458002 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 11 |
Hb_052113_020 |
0.1905346556 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 12 |
Hb_116349_090 |
0.192906722 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 13 |
Hb_078901_010 |
0.196193138 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_000805_150 |
0.1975276631 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000856_410 |
0.197632744 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_019516_050 |
0.1977682189 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |
| 17 |
Hb_000434_050 |
0.1999079745 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 18 |
Hb_125884_010 |
0.2008752847 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Jatropha curcas] |
| 19 |
Hb_029238_020 |
0.2022867059 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 20 |
Hb_001956_190 |
0.2030186372 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |