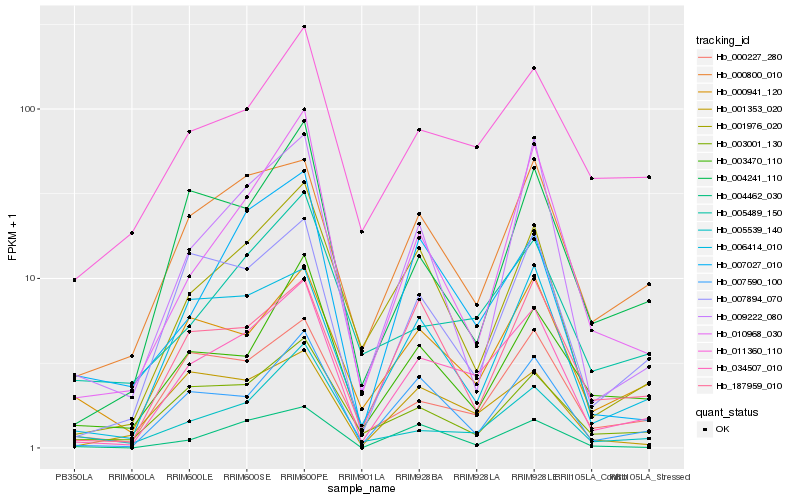
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034507_010 |
0.0 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 2 |
Hb_007590_100 |
0.1454855319 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 3 |
Hb_001976_020 |
0.156818783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000227_280 |
0.1569546979 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 5 |
Hb_007894_070 |
0.1589050781 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 6 |
Hb_006414_010 |
0.1616022439 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 7 |
Hb_000800_010 |
0.1625590901 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 8 |
Hb_011360_110 |
0.1631500875 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 9 |
Hb_010968_030 |
0.1652803735 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 10 |
Hb_003001_130 |
0.1689821011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004241_110 |
0.1697000579 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 12 |
Hb_000941_120 |
0.1704951278 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 13 |
Hb_001353_020 |
0.171583835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005489_150 |
0.1744812478 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 15 |
Hb_007027_010 |
0.1759346982 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 16 |
Hb_003470_110 |
0.1765141271 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_009222_080 |
0.1788060372 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 18 |
Hb_004462_030 |
0.1791432255 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 19 |
Hb_005539_140 |
0.1803184792 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 20 |
Hb_187959_010 |
0.1818349947 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |