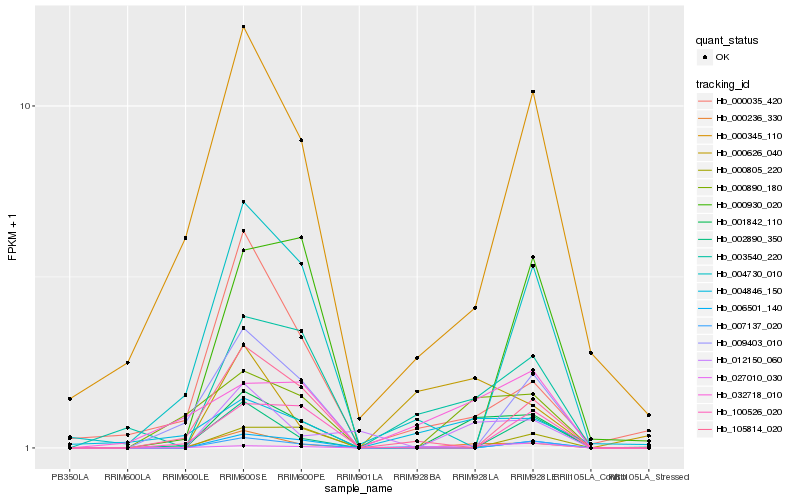
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001842_110 |
0.0 |
transcription factor |
TF Family: ERF |
CBF-like transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000236_330 |
0.198231189 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000890_180 |
0.2497489055 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 4 |
Hb_012150_060 |
0.2689567634 |
- |
- |
- |
| 5 |
Hb_003540_220 |
0.2704780509 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 6 |
Hb_006501_140 |
0.3324382846 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 7 |
Hb_032718_010 |
0.3351304983 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_027010_030 |
0.3371511971 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_000345_110 |
0.3497820507 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 10 |
Hb_000035_420 |
0.3537398397 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 11 |
Hb_004846_150 |
0.3546005286 |
- |
- |
hypothetical protein JCGZ_26968 [Jatropha curcas] |
| 12 |
Hb_000805_220 |
0.3579435102 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_007137_020 |
0.3660017464 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 14 |
Hb_105814_020 |
0.3702019537 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 15 |
Hb_002890_350 |
0.3712237131 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_009403_010 |
0.3713863147 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 17 |
Hb_000626_040 |
0.3714073312 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 18 |
Hb_100526_020 |
0.3729076296 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 19 |
Hb_000930_020 |
0.3761759268 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 20 |
Hb_004730_010 |
0.3789270642 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |