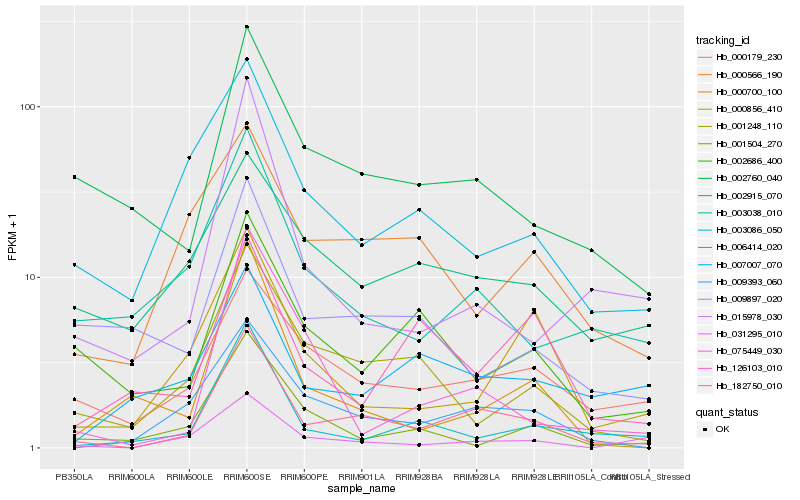
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182750_010 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a0001602mg, partial [Erythranthe guttata] |
| 2 |
Hb_009393_060 |
0.2062932686 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 3 |
Hb_002760_040 |
0.2317221188 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 4 |
Hb_000179_230 |
0.2336266473 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 5 |
Hb_001248_110 |
0.2361089244 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_003086_050 |
0.2395786299 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_031295_010 |
0.2428853127 |
- |
- |
hypothetical protein JCGZ_19530 [Jatropha curcas] |
| 8 |
Hb_003038_010 |
0.2475400507 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000700_100 |
0.2534375354 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 10 |
Hb_015978_030 |
0.2547747439 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 11 |
Hb_002686_400 |
0.2563642848 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 12 |
Hb_006414_020 |
0.2579222905 |
transcription factor |
TF Family: MYB |
- |
| 13 |
Hb_000856_410 |
0.2599788077 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_000566_190 |
0.2602077079 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_009897_020 |
0.2648971187 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_075449_030 |
0.2651055025 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 17 |
Hb_126103_010 |
0.2699899797 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 18 |
Hb_007007_070 |
0.2715891461 |
transcription factor |
TF Family: TCP |
hypothetical protein MIMGU_mgv1a022855mg, partial [Erythranthe guttata] |
| 19 |
Hb_002915_070 |
0.2720933461 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 20 |
Hb_001504_270 |
0.2749153104 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |