| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
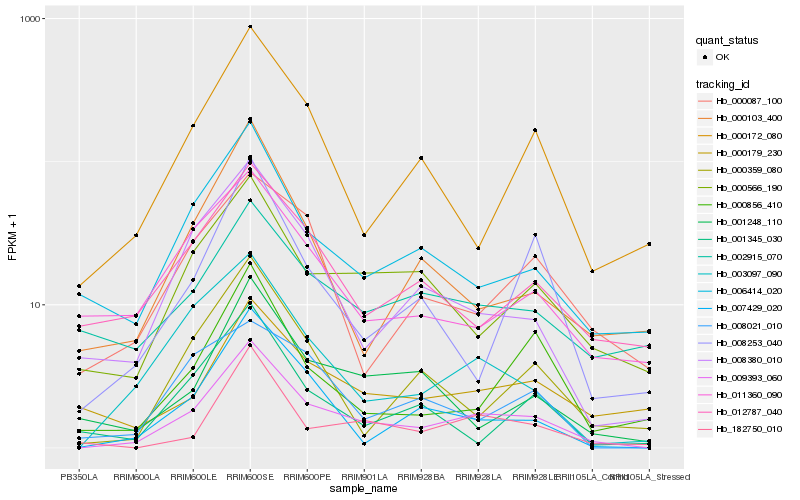
Hb_009393_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 2 |
Hb_003097_090 |
0.1880624218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007429_020 |
0.1948680189 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000566_190 |
0.1970183971 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_006414_020 |
0.1995965435 |
transcription factor |
TF Family: MYB |
- |
| 6 |
Hb_008380_010 |
0.200241345 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 7 |
Hb_182750_010 |
0.2062932686 |
- |
- |
hypothetical protein MIMGU_mgv1a0001602mg, partial [Erythranthe guttata] |
| 8 |
Hb_000172_080 |
0.2080077466 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 9 |
Hb_001248_110 |
0.2130283767 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_000359_080 |
0.2131806664 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 11 |
Hb_000103_400 |
0.2191339745 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012787_040 |
0.2203791647 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 13 |
Hb_008253_040 |
0.2215303097 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 14 |
Hb_000179_230 |
0.2225367923 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 15 |
Hb_000856_410 |
0.2228847023 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_000087_100 |
0.2286882027 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 17 |
Hb_002915_070 |
0.2320548174 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 18 |
Hb_011360_090 |
0.2322998995 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 19 |
Hb_008021_010 |
0.232609335 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 20 |
Hb_001345_030 |
0.2337332813 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |