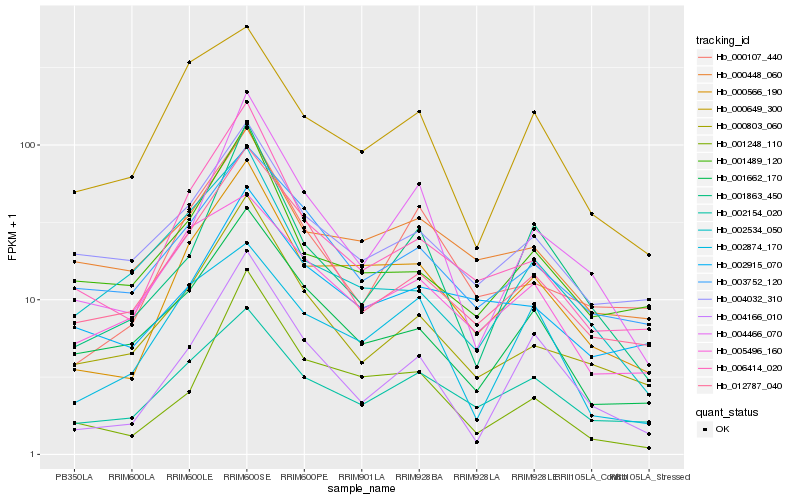
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000566_190 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_006414_020 |
0.1264312717 |
transcription factor |
TF Family: MYB |
- |
| 3 |
Hb_002154_020 |
0.1356719578 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000448_060 |
0.1459669905 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 5 |
Hb_000107_440 |
0.1478659122 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 6 |
Hb_001248_110 |
0.1521015299 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 7 |
Hb_012787_040 |
0.1530057987 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001489_120 |
0.1538507601 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000803_060 |
0.1543943799 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 10 |
Hb_001662_170 |
0.1549356231 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 11 |
Hb_000649_300 |
0.1560228863 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 12 |
Hb_003752_120 |
0.1572133449 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004032_310 |
0.1577049206 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 14 |
Hb_002915_070 |
0.1591667253 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 15 |
Hb_004166_010 |
0.1627691452 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 16 |
Hb_002874_170 |
0.1668366109 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_005496_160 |
0.1670521285 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001863_450 |
0.1671832942 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 19 |
Hb_004466_070 |
0.1675001354 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 20 |
Hb_002534_050 |
0.1717597914 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |