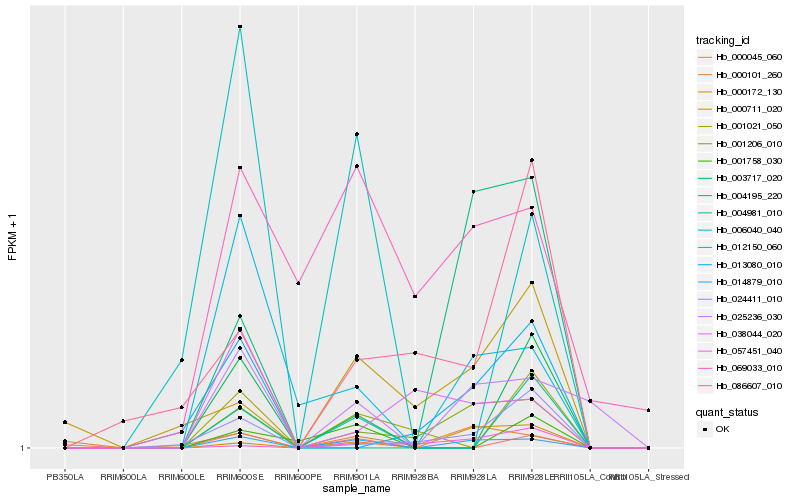
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014879_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_001206_010 |
0.2398476372 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 3 |
Hb_012150_060 |
0.2667201875 |
- |
- |
- |
| 4 |
Hb_025236_030 |
0.2809563563 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 5 |
Hb_000172_130 |
0.3068435976 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 6 |
Hb_000101_260 |
0.3150963332 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_004981_010 |
0.382637034 |
- |
- |
- |
| 8 |
Hb_000711_020 |
0.3835916363 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 9 |
Hb_069033_010 |
0.3857896059 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 10 |
Hb_024411_010 |
0.3871754621 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_003717_020 |
0.3883453464 |
- |
- |
- |
| 12 |
Hb_000045_060 |
0.3914116377 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_006040_040 |
0.39168772 |
- |
- |
Putative mitochondrial protein [Glycine soja] |
| 14 |
Hb_038044_020 |
0.3981697141 |
- |
- |
- |
| 15 |
Hb_004195_220 |
0.4052743516 |
- |
- |
hypothetical protein PRUPE_ppa005288mg [Prunus persica] |
| 16 |
Hb_057451_040 |
0.408885706 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_013080_010 |
0.41508109 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_086607_010 |
0.4153333499 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_001021_050 |
0.416494448 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 20 |
Hb_001758_030 |
0.4172596907 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |