| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
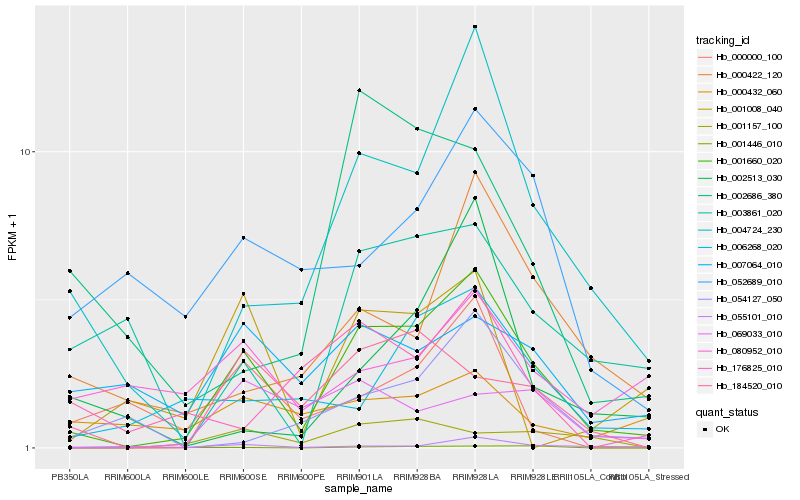
Hb_001660_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 2 |
Hb_004724_230 |
0.2143098846 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 3 |
Hb_184520_010 |
0.2695077485 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 4 |
Hb_006268_020 |
0.2816348138 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 5 |
Hb_000422_120 |
0.2845613777 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 6 |
Hb_055101_010 |
0.2847602977 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_007064_010 |
0.2872617202 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 8 |
Hb_052689_010 |
0.2890009325 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 9 |
Hb_069033_010 |
0.2931166057 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 10 |
Hb_001157_100 |
0.2935918804 |
- |
- |
PREDICTED: uncharacterized protein LOC104887185 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_001446_010 |
0.3005346678 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_054127_050 |
0.3046568533 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003861_020 |
0.3058986032 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like isoform X2 [Nelumbo nucifera] |
| 14 |
Hb_000000_100 |
0.3065958539 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 15 |
Hb_002686_380 |
0.3085665812 |
- |
- |
- |
| 16 |
Hb_176825_010 |
0.3125536236 |
- |
- |
PREDICTED: uncharacterized protein LOC105630857 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002513_030 |
0.3128308777 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 18 |
Hb_080952_010 |
0.3129737436 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 19 |
Hb_001008_040 |
0.3135922465 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 20 |
Hb_000432_060 |
0.3138485731 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |