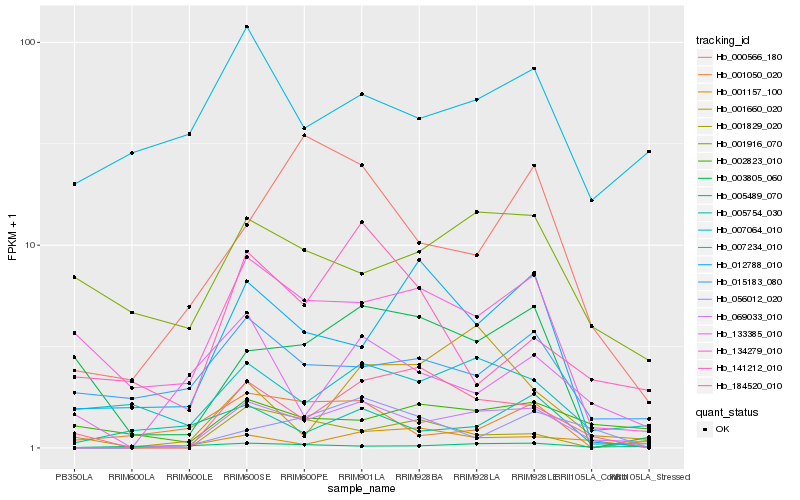
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069033_010 |
0.0 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 2 |
Hb_184520_010 |
0.2617823674 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_001157_100 |
0.2618894511 |
- |
- |
PREDICTED: uncharacterized protein LOC104887185 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_007064_010 |
0.2762954379 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 5 |
Hb_001829_020 |
0.2788301053 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 6 |
Hb_000566_180 |
0.2789368839 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 7 |
Hb_056012_020 |
0.2825230298 |
- |
- |
- |
| 8 |
Hb_134279_010 |
0.2843458477 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 9 |
Hb_133385_010 |
0.2898121914 |
- |
- |
- |
| 10 |
Hb_001660_020 |
0.2931166057 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 11 |
Hb_003805_060 |
0.2981926169 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_007234_010 |
0.2998491613 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 13 |
Hb_002823_010 |
0.3004800415 |
- |
- |
- |
| 14 |
Hb_015183_080 |
0.3024298385 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_001916_070 |
0.3029587738 |
- |
- |
PREDICTED: phytochrome C [Jatropha curcas] |
| 16 |
Hb_005754_030 |
0.3060289167 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 17 |
Hb_001050_020 |
0.3064975637 |
- |
- |
- |
| 18 |
Hb_141212_010 |
0.3067613902 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_005489_070 |
0.3069540027 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 20 |
Hb_012788_010 |
0.3098121553 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |