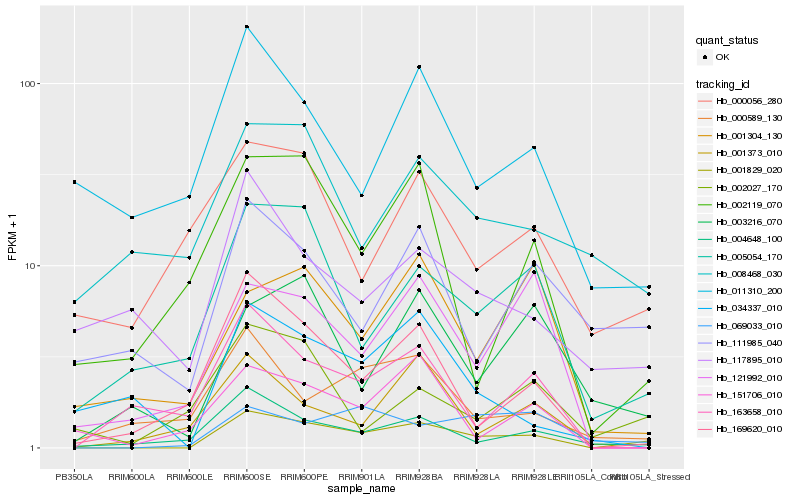
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001829_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 2 |
Hb_004648_100 |
0.256874621 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 3 |
Hb_005054_170 |
0.260197926 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_169620_010 |
0.2649114298 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 5 |
Hb_069033_010 |
0.2788301053 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 6 |
Hb_003216_070 |
0.2834360241 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 7 |
Hb_002119_070 |
0.2858948992 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_111985_040 |
0.2886328217 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 9 |
Hb_163658_010 |
0.2911174751 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_121992_010 |
0.2921125674 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 11 |
Hb_001373_010 |
0.292966821 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_151706_010 |
0.2980814826 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 13 |
Hb_000589_130 |
0.2986623997 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000056_280 |
0.2987447144 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 15 |
Hb_034337_010 |
0.3020313246 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 16 |
Hb_002027_170 |
0.3026977774 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 17 |
Hb_117895_010 |
0.3044159905 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 18 |
Hb_011310_200 |
0.3048640289 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 19 |
Hb_001304_130 |
0.3051686419 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 20 |
Hb_008468_030 |
0.3054086369 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |