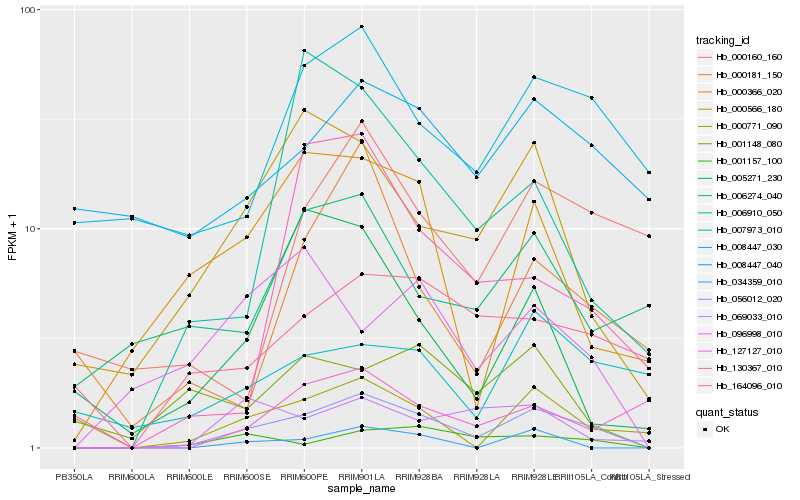
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056012_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000566_180 |
0.2519569668 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 3 |
Hb_001157_100 |
0.2629023243 |
- |
- |
PREDICTED: uncharacterized protein LOC104887185 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000366_020 |
0.2693227365 |
- |
- |
- |
| 5 |
Hb_008447_030 |
0.2781965898 |
- |
- |
- |
| 6 |
Hb_069033_010 |
0.2825230298 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 7 |
Hb_034359_010 |
0.2865515614 |
- |
- |
- |
| 8 |
Hb_164096_010 |
0.2944123902 |
- |
- |
hypothetical protein L484_021244 [Morus notabilis] |
| 9 |
Hb_005271_230 |
0.2977049278 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 10 |
Hb_000771_090 |
0.3000422718 |
- |
- |
hypothetical protein B456_012G003500 [Gossypium raimondii] |
| 11 |
Hb_006910_050 |
0.3001976805 |
- |
- |
hypothetical protein CISIN_1g0480481mg, partial [Citrus sinensis] |
| 12 |
Hb_001148_080 |
0.3002018004 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 13 |
Hb_096998_010 |
0.3008441069 |
- |
- |
hypothetical protein JCGZ_07944 [Jatropha curcas] |
| 14 |
Hb_000160_160 |
0.3030191834 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000181_150 |
0.30451744 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 16 |
Hb_127127_010 |
0.3046483101 |
- |
- |
- |
| 17 |
Hb_008447_040 |
0.3054022637 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 18 |
Hb_130367_010 |
0.3062220994 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 19 |
Hb_006274_040 |
0.307738969 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_007973_010 |
0.3079105956 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |