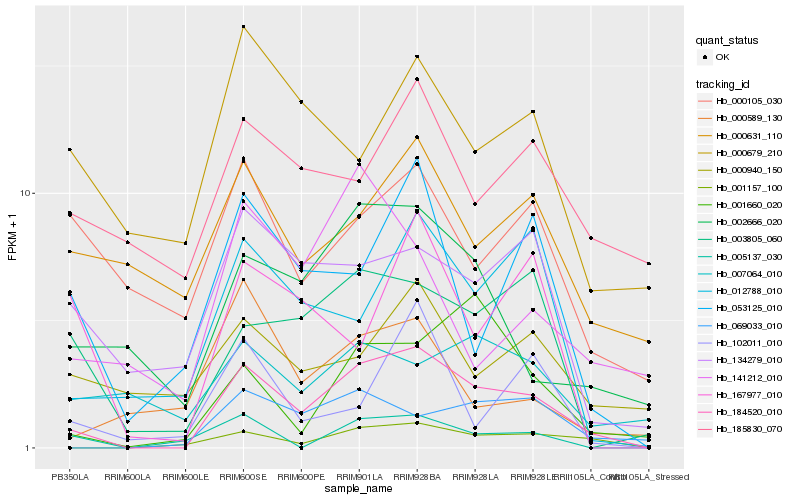
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_184520_010 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_001157_100 |
0.221158517 |
- |
- |
PREDICTED: uncharacterized protein LOC104887185 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_002666_020 |
0.2539657073 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 4 |
Hb_003805_060 |
0.2582446005 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_012788_010 |
0.2583790174 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_069033_010 |
0.2617823674 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 7 |
Hb_053125_010 |
0.2646781143 |
- |
- |
PREDICTED: uncharacterized protein LOC104881508 [Vitis vinifera] |
| 8 |
Hb_001660_020 |
0.2695077485 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 9 |
Hb_134279_010 |
0.2733200966 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 10 |
Hb_005137_030 |
0.281697464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_167977_010 |
0.2923668618 |
- |
- |
hypothetical protein POPTR_0185s002102g, partial [Populus trichocarpa] |
| 12 |
Hb_000631_110 |
0.2944925595 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 13 |
Hb_000589_130 |
0.2979886312 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000679_210 |
0.2985632032 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 15 |
Hb_000940_150 |
0.2990499261 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 16 |
Hb_102011_010 |
0.2993450597 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_000105_030 |
0.300038002 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 18 |
Hb_007064_010 |
0.3006541712 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 19 |
Hb_141212_010 |
0.301588532 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 20 |
Hb_185830_070 |
0.3025076644 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |