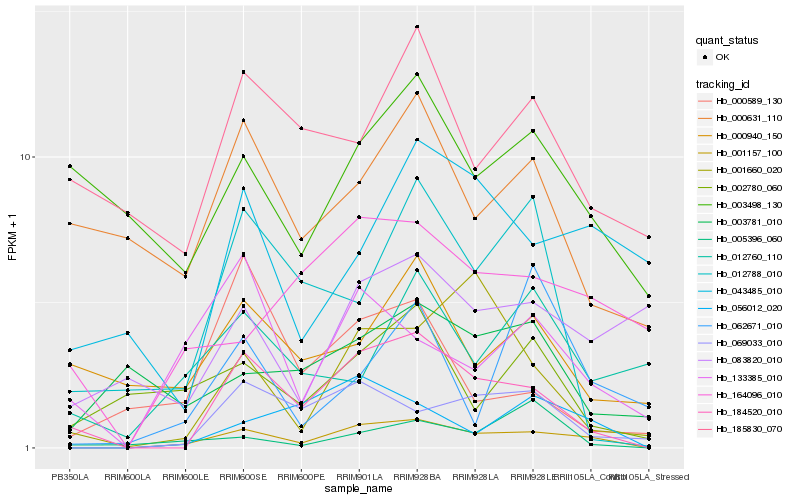
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104887185 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_184520_010 |
0.221158517 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_069033_010 |
0.2618894511 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 4 |
Hb_056012_020 |
0.2629023243 |
- |
- |
- |
| 5 |
Hb_133385_010 |
0.2904161172 |
- |
- |
- |
| 6 |
Hb_001660_020 |
0.2935918804 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 7 |
Hb_164096_010 |
0.2939535899 |
- |
- |
hypothetical protein L484_021244 [Morus notabilis] |
| 8 |
Hb_002780_060 |
0.3009260686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_012788_010 |
0.3090563037 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_043485_010 |
0.3107564987 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_083820_010 |
0.3121852654 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 12 |
Hb_003781_010 |
0.3132057297 |
- |
- |
PREDICTED: histone H2AX [Eucalyptus grandis] |
| 13 |
Hb_012760_110 |
0.3194994333 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000940_150 |
0.3240362844 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 15 |
Hb_000631_110 |
0.3245986644 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_000589_130 |
0.3273663584 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_005396_060 |
0.3277485495 |
- |
- |
- |
| 18 |
Hb_185830_070 |
0.3280008559 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_062671_010 |
0.3281322825 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003498_130 |
0.3288235058 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |