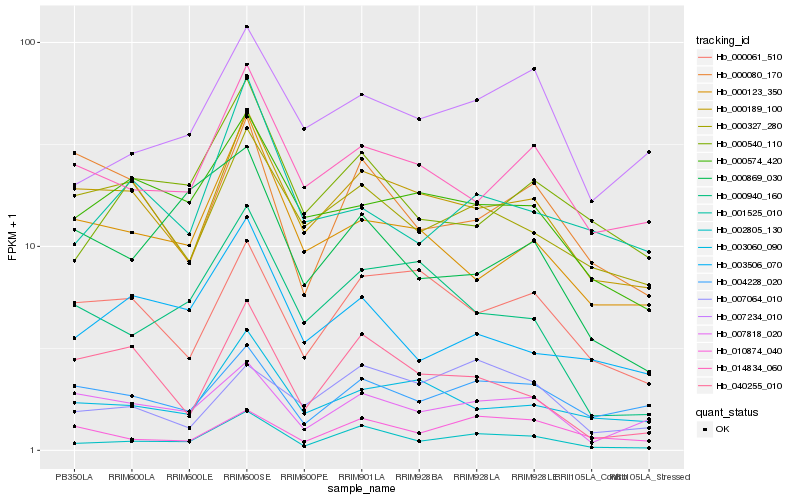
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002805_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000189_100 |
0.1749877338 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 3 |
Hb_000540_110 |
0.1803359047 |
- |
- |
hypothetical protein PRUPE_ppa000777mg [Prunus persica] |
| 4 |
Hb_007234_010 |
0.185133052 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 5 |
Hb_000940_160 |
0.1868733843 |
- |
- |
hypothetical protein VITISV_017596 [Vitis vinifera] |
| 6 |
Hb_004228_020 |
0.1884614307 |
- |
- |
hypothetical protein JCGZ_21866 [Jatropha curcas] |
| 7 |
Hb_007818_020 |
0.1884752358 |
- |
- |
predicted protein [Physcomitrella patens] |
| 8 |
Hb_014834_060 |
0.189406828 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 9 |
Hb_010874_040 |
0.192360223 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein At3g13880 [Jatropha curcas] |
| 10 |
Hb_040255_010 |
0.1932786476 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 11 |
Hb_003506_070 |
0.1961591668 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003060_090 |
0.1966517228 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 13 |
Hb_000061_510 |
0.200132738 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 14 |
Hb_007064_010 |
0.2015142204 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 15 |
Hb_000574_420 |
0.2032692527 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 16 |
Hb_000123_350 |
0.203595074 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000869_030 |
0.2053010887 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001525_010 |
0.2057053669 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 19 |
Hb_000327_280 |
0.2068490926 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000080_170 |
0.2071581493 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |