| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
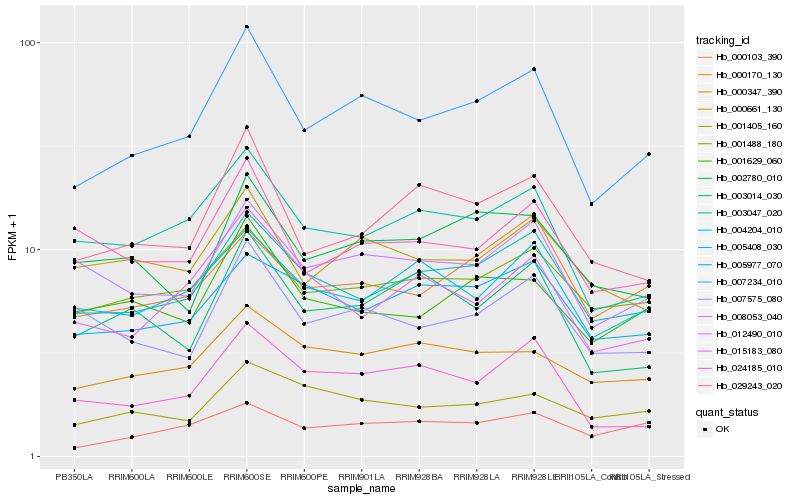
Hb_007234_010 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 2 |
Hb_004204_010 |
0.0956877772 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 3 |
Hb_029243_020 |
0.1057565444 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 4 |
Hb_003047_020 |
0.1065624333 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_008053_040 |
0.1080582734 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 6 |
Hb_015183_080 |
0.1084817572 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001488_180 |
0.1085855479 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000347_390 |
0.1147618594 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 9 |
Hb_000661_130 |
0.114873458 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 10 |
Hb_001629_060 |
0.1172386074 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003014_030 |
0.1175219939 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |
| 12 |
Hb_007575_080 |
0.1175222119 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 13 |
Hb_005977_070 |
0.1189128488 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002780_010 |
0.1194155234 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 15 |
Hb_000170_130 |
0.1195030153 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 16 |
Hb_000103_390 |
0.119665781 |
- |
- |
Pentatricopeptide repeat superfamily protein [Theobroma cacao] |
| 17 |
Hb_005408_030 |
0.1204096172 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 18 |
Hb_001405_160 |
0.1220860369 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 19 |
Hb_012490_010 |
0.122572627 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 20 |
Hb_024185_010 |
0.1231811062 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |