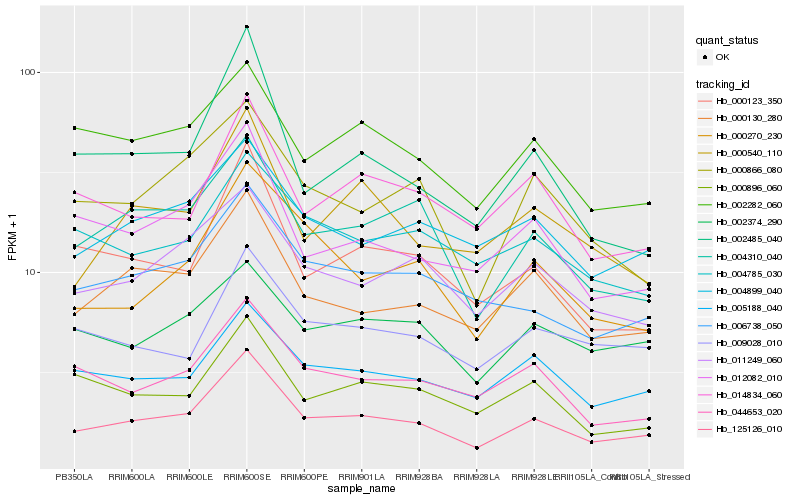
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_125126_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 2 |
Hb_000130_280 |
0.0823799308 |
- |
- |
PREDICTED: transcription factor GTE3, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_006738_050 |
0.0929057147 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 4 |
Hb_004310_040 |
0.0938523781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011249_060 |
0.0957295054 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 6 |
Hb_005188_040 |
0.0969158137 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 7 |
Hb_000123_350 |
0.0990369602 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_004899_040 |
0.0995124178 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 9 |
Hb_012082_010 |
0.0997602947 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_014834_060 |
0.1048750047 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 11 |
Hb_044653_020 |
0.1048829307 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 12 |
Hb_000270_230 |
0.1066004737 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 13 |
Hb_000540_110 |
0.1078400128 |
- |
- |
hypothetical protein PRUPE_ppa000777mg [Prunus persica] |
| 14 |
Hb_002282_060 |
0.1078499671 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_009028_010 |
0.1095522272 |
- |
- |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 16 |
Hb_000866_080 |
0.1113013049 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002485_040 |
0.1116764966 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 18 |
Hb_002374_290 |
0.1116835666 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000896_060 |
0.1122777255 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_004785_030 |
0.1130425298 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |