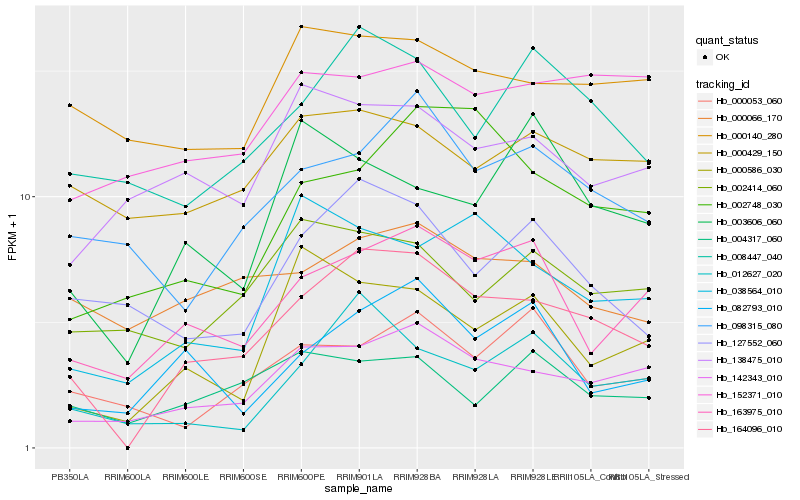
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164096_010 |
0.0 |
- |
- |
hypothetical protein L484_021244 [Morus notabilis] |
| 2 |
Hb_142343_010 |
0.1500555042 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_163975_010 |
0.1728718874 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_008447_040 |
0.1847730508 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 5 |
Hb_038564_010 |
0.1886669026 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_012627_020 |
0.1909041147 |
- |
- |
PHYTOCHROME AND FLOWERING TIME 1 family protein [Populus trichocarpa] |
| 7 |
Hb_098315_080 |
0.192011753 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_002748_030 |
0.1926292602 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 9 |
Hb_000586_030 |
0.1930029191 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 10 |
Hb_082793_010 |
0.1946916143 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_127552_060 |
0.1956793877 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_000066_170 |
0.1994458995 |
- |
- |
PREDICTED: phosphatidylinositol-3-phosphatase myotubularin-1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004317_060 |
0.2036487027 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 14 |
Hb_002414_060 |
0.2048311376 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 15 |
Hb_000429_150 |
0.2053354574 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 16 |
Hb_152371_010 |
0.2064237473 |
- |
- |
RNA-binding KH domain-containing protein [Theobroma cacao] |
| 17 |
Hb_003606_060 |
0.2074244592 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 18 |
Hb_000140_280 |
0.207860198 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 19 |
Hb_138475_010 |
0.2094533762 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Populus euphratica] |
| 20 |
Hb_000053_060 |
0.2102253582 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |