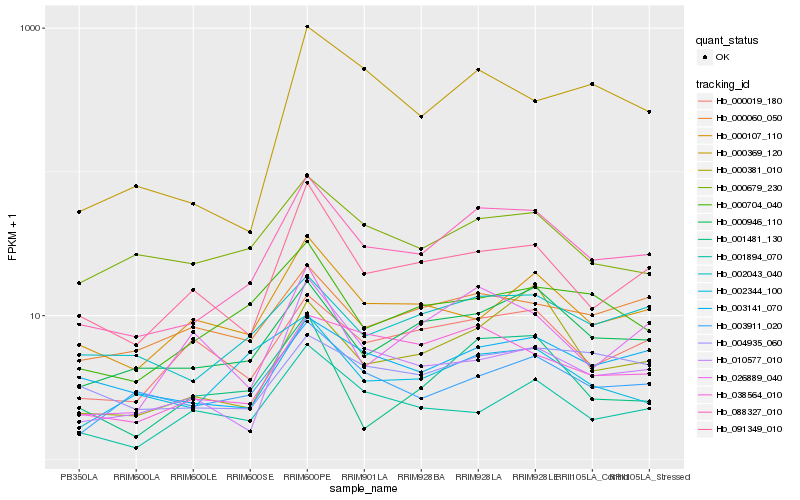
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088327_010 |
0.0 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 2 |
Hb_000946_110 |
0.1311324745 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 3 |
Hb_091349_010 |
0.1406939326 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 4 |
Hb_038564_010 |
0.1413235904 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002043_040 |
0.1428131056 |
- |
- |
PREDICTED: uncharacterized protein LOC105631509 [Jatropha curcas] |
| 6 |
Hb_000704_040 |
0.1467549162 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 7 |
Hb_000381_010 |
0.1500351511 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_003911_020 |
0.1503012378 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 9 |
Hb_003141_070 |
0.1530809475 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 10 |
Hb_000019_180 |
0.1567782066 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 11 |
Hb_000107_110 |
0.1613271148 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000679_230 |
0.1647362538 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 13 |
Hb_000369_120 |
0.1663724209 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 14 |
Hb_010577_010 |
0.1678667822 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 15 |
Hb_001894_070 |
0.1682521202 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004935_060 |
0.1687911051 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_002344_100 |
0.1688288157 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 18 |
Hb_001481_130 |
0.1698312986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000060_050 |
0.1704409457 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 20 |
Hb_026889_040 |
0.1708570038 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |