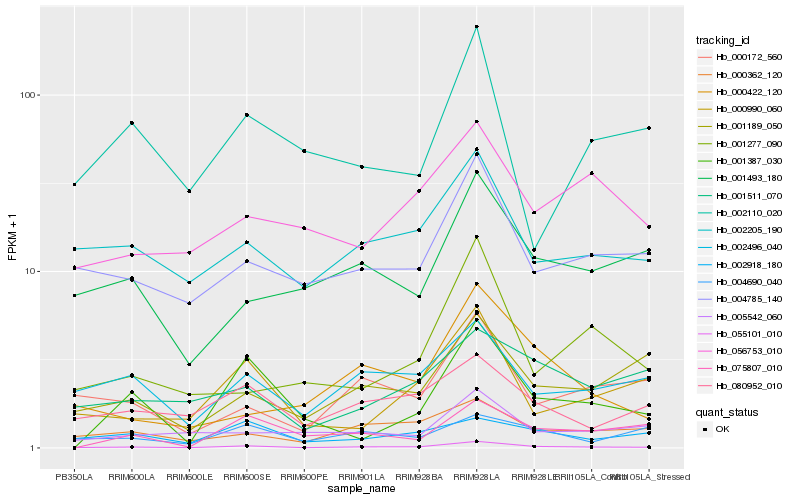
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055101_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_004785_140 |
0.1888360889 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000990_060 |
0.1901017843 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 4 |
Hb_002205_190 |
0.1947147468 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 5 |
Hb_080952_010 |
0.194772702 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 6 |
Hb_000362_120 |
0.2006810128 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 7 |
Hb_002496_040 |
0.2017156485 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001189_050 |
0.204440478 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 9 |
Hb_000172_560 |
0.2064293047 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 10 |
Hb_005542_060 |
0.207436602 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000422_120 |
0.2100436458 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 12 |
Hb_001493_180 |
0.2149096963 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 13 |
Hb_001277_090 |
0.2160367633 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 14 |
Hb_001387_030 |
0.2202833876 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 15 |
Hb_002918_180 |
0.2232348487 |
- |
- |
hypothetical protein POPTR_0004s04740g [Populus trichocarpa] |
| 16 |
Hb_004690_040 |
0.2233498351 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 17 |
Hb_001511_070 |
0.2252144884 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 18 |
Hb_056753_010 |
0.2264205986 |
- |
- |
PREDICTED: L-galactono-1,4-lactone dehydrogenase, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_002110_020 |
0.2304473564 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 20 |
Hb_075807_010 |
0.2315782742 |
- |
- |
PREDICTED: uncharacterized protein LOC105631775 [Jatropha curcas] |