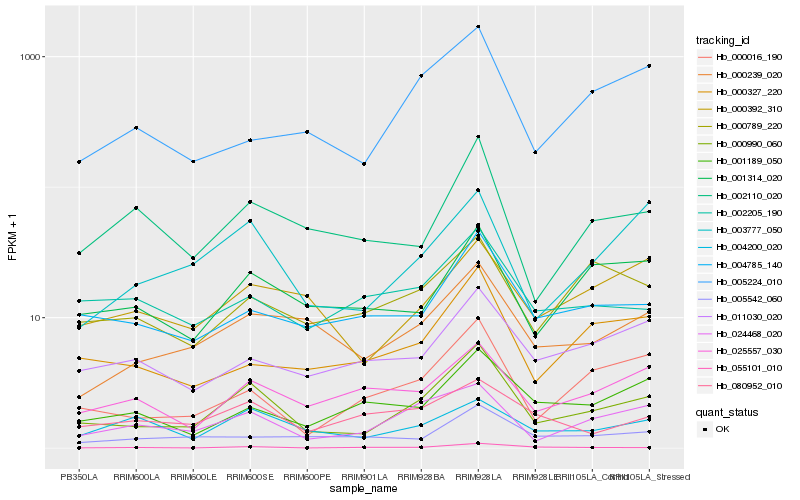
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000990_060 |
0.0 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 2 |
Hb_024468_020 |
0.1672592339 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 3 |
Hb_000016_190 |
0.1785249697 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 4 |
Hb_003777_050 |
0.1794901087 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 5 |
Hb_000239_020 |
0.1891528719 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 6 |
Hb_055101_010 |
0.1901017843 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_004785_140 |
0.1924287294 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000327_220 |
0.1956690885 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 9 |
Hb_002110_020 |
0.1992902878 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 10 |
Hb_001314_020 |
0.1996454005 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_005224_010 |
0.2036538752 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 12 |
Hb_025557_030 |
0.204311206 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 13 |
Hb_011030_020 |
0.2064374205 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 14 |
Hb_000392_310 |
0.2072144326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004200_020 |
0.2094582858 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 16 |
Hb_001189_050 |
0.2124924224 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 17 |
Hb_000789_220 |
0.2126974262 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 18 |
Hb_002205_190 |
0.2150036535 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 19 |
Hb_005542_060 |
0.2154586104 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_080952_010 |
0.2166372238 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |