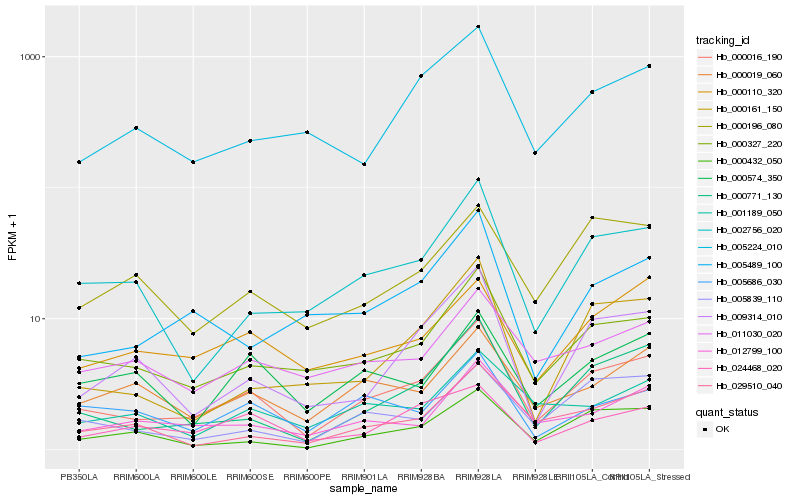
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_190 |
0.0 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 2 |
Hb_000771_130 |
0.1143807152 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_000327_220 |
0.1218899288 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 4 |
Hb_002756_020 |
0.1450536368 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 5 |
Hb_000432_050 |
0.1479787968 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 6 |
Hb_012799_100 |
0.1509339162 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 7 |
Hb_009314_010 |
0.1565791035 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 8 |
Hb_005224_010 |
0.1568623048 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 9 |
Hb_000019_060 |
0.1592969943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial [Jatropha curcas] |
| 10 |
Hb_005839_110 |
0.1606513496 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Populus euphratica] |
| 11 |
Hb_029510_040 |
0.1612546323 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 12 |
Hb_024468_020 |
0.1612749341 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_000110_320 |
0.1618870835 |
- |
- |
PREDICTED: ceramide-1-phosphate transfer protein [Jatropha curcas] |
| 14 |
Hb_000161_150 |
0.1618978781 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 15 |
Hb_005489_100 |
0.1642509122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011030_020 |
0.1664511179 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 17 |
Hb_000574_350 |
0.1678070887 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 18 |
Hb_005686_030 |
0.1679941771 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_001189_050 |
0.1693452518 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 20 |
Hb_000196_080 |
0.1709179153 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |