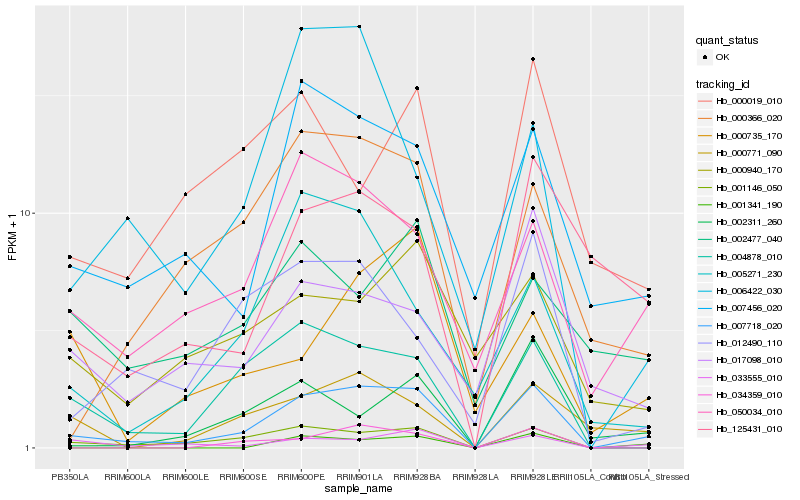
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007718_020 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 2 |
Hb_001146_050 |
0.1958106007 |
- |
- |
- |
| 3 |
Hb_004878_010 |
0.2166255523 |
- |
- |
- |
| 4 |
Hb_033555_010 |
0.2249702199 |
- |
- |
- |
| 5 |
Hb_050034_010 |
0.2289382577 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 6 |
Hb_007456_020 |
0.2290797787 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_000771_090 |
0.2418019749 |
- |
- |
hypothetical protein B456_012G003500 [Gossypium raimondii] |
| 8 |
Hb_000366_020 |
0.2446410126 |
- |
- |
- |
| 9 |
Hb_125431_010 |
0.2522009182 |
- |
- |
- |
| 10 |
Hb_012490_110 |
0.2536054953 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 11 |
Hb_034359_010 |
0.2588668129 |
- |
- |
- |
| 12 |
Hb_017098_010 |
0.2666375959 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_001341_190 |
0.266926466 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002311_260 |
0.2676659961 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 15 |
Hb_000940_170 |
0.2700445424 |
- |
- |
hypothetical protein VITISV_029834 [Vitis vinifera] |
| 16 |
Hb_005271_230 |
0.2761426324 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 17 |
Hb_000019_010 |
0.2763444357 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |
| 18 |
Hb_000735_170 |
0.2781164594 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 19 |
Hb_006422_030 |
0.2784195182 |
- |
- |
- |
| 20 |
Hb_002477_040 |
0.2812914917 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein isoform 2 [Theobroma cacao] |