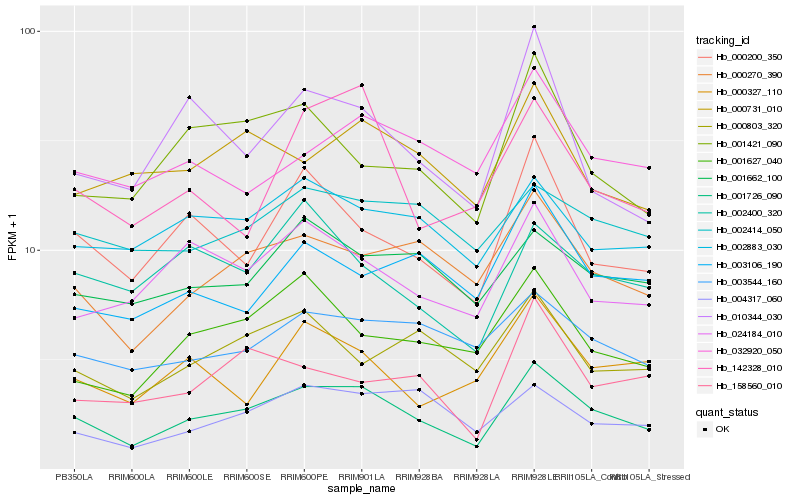
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001726_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000270_390 |
0.1236021723 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 3 |
Hb_000200_350 |
0.1289342316 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 4 |
Hb_024184_010 |
0.1378161304 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 5 |
Hb_001627_040 |
0.1381798368 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001421_090 |
0.1384910778 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 7 |
Hb_002883_030 |
0.1404796465 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 8 |
Hb_004317_060 |
0.1406766623 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_003544_160 |
0.1408340636 |
- |
- |
- |
| 10 |
Hb_001662_100 |
0.1447731989 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 11 |
Hb_010344_030 |
0.1450614466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000731_010 |
0.1459495419 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_142328_010 |
0.1476743351 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002400_320 |
0.1482511036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000803_320 |
0.1485129087 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 16 |
Hb_032920_050 |
0.149190475 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002414_050 |
0.1514611456 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 18 |
Hb_003106_190 |
0.1524998697 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 19 |
Hb_158560_010 |
0.1530134235 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 20 |
Hb_000327_110 |
0.1540278136 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |