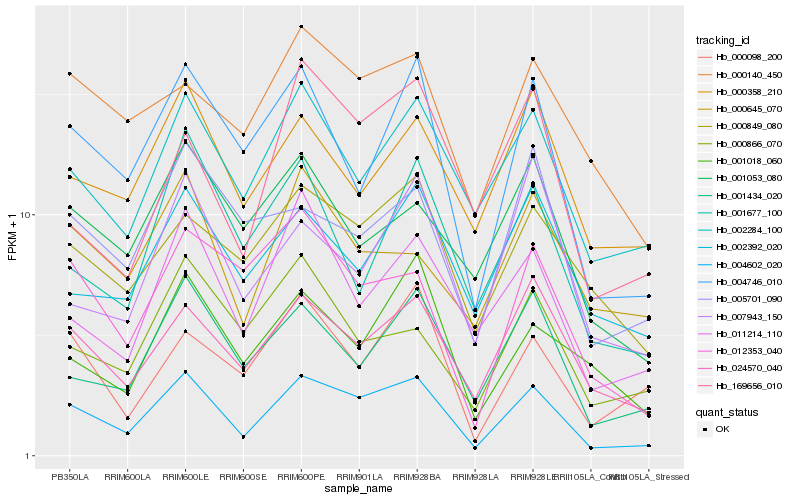
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004602_020 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: N-lysine methyltransferase setd6 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004746_010 |
0.1370275246 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 3 |
Hb_024570_040 |
0.1401842179 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 4 |
Hb_001434_020 |
0.1452575682 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 5 |
Hb_002284_100 |
0.1510595012 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 6 |
Hb_011214_110 |
0.1531249999 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 7 |
Hb_000849_080 |
0.162377817 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000098_200 |
0.1630454974 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like isoform X2 [Populus euphratica] |
| 9 |
Hb_012353_040 |
0.1667605833 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_001018_060 |
0.1670331216 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 11 |
Hb_000358_210 |
0.1710073092 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 12 |
Hb_169656_010 |
0.1710686403 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 13 |
Hb_005701_090 |
0.1712626376 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001677_100 |
0.1732623193 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 15 |
Hb_002392_020 |
0.1733798539 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 16 |
Hb_000140_450 |
0.1736435667 |
- |
- |
PREDICTED: calreticulin-3-like [Populus euphratica] |
| 17 |
Hb_000645_070 |
0.1742555414 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_007943_150 |
0.175106848 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000866_070 |
0.1751867497 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 20 |
Hb_001053_080 |
0.1752810616 |
- |
- |
OsCesA3 protein [Morus notabilis] |