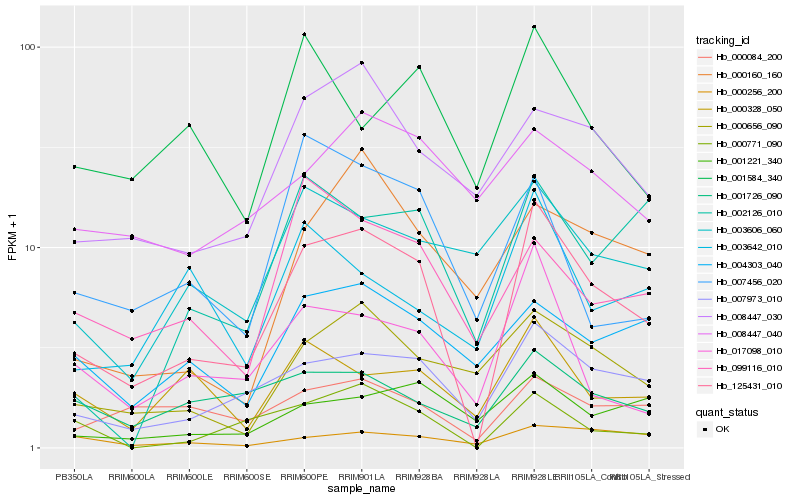
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_125431_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_007973_010 |
0.1667982193 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000771_090 |
0.2071180397 |
- |
- |
hypothetical protein B456_012G003500 [Gossypium raimondii] |
| 4 |
Hb_017098_010 |
0.212489255 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000656_090 |
0.2204673339 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 6 |
Hb_008447_030 |
0.2205847483 |
- |
- |
- |
| 7 |
Hb_000084_200 |
0.2227306316 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001726_090 |
0.229903482 |
- |
- |
- |
| 9 |
Hb_000328_050 |
0.2321450066 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004303_040 |
0.2355296214 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001221_340 |
0.235832464 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 12 |
Hb_099116_010 |
0.2371902982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003606_060 |
0.2374519637 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 14 |
Hb_007456_020 |
0.2390101582 |
- |
- |
unknown [Lotus japonicus] |
| 15 |
Hb_001584_340 |
0.2402448924 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 16 |
Hb_002126_010 |
0.240905995 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 17 |
Hb_000256_200 |
0.2415762246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000160_160 |
0.2418306343 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_008447_040 |
0.2456309344 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 20 |
Hb_003642_010 |
0.2460955717 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |