| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
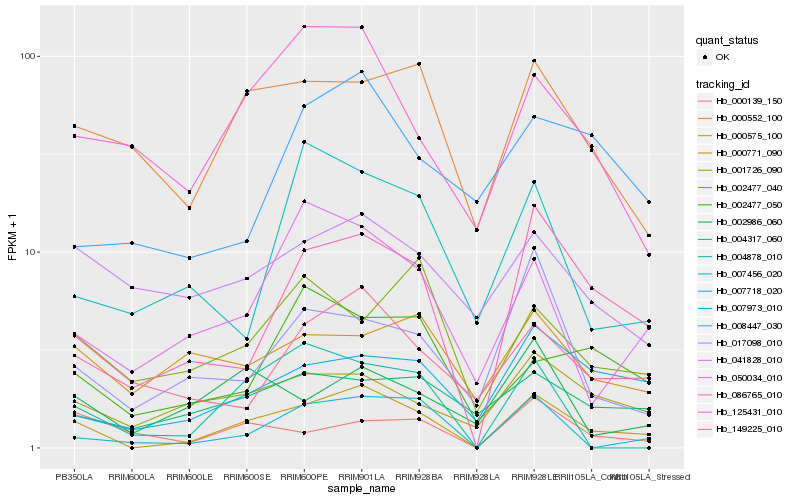
Hb_000771_090 |
0.0 |
- |
- |
hypothetical protein B456_012G003500 [Gossypium raimondii] |
| 2 |
Hb_125431_010 |
0.2071180397 |
- |
- |
- |
| 3 |
Hb_007718_020 |
0.2418019749 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 4 |
Hb_007973_010 |
0.2434733489 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001726_090 |
0.2495758383 |
- |
- |
- |
| 6 |
Hb_050034_010 |
0.2508959706 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 7 |
Hb_149225_010 |
0.2536094329 |
- |
- |
DNA-directed RNA polymerase II subunit rpb4, putative isoform 4, partial [Theobroma cacao] |
| 8 |
Hb_000552_100 |
0.2539201959 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 9 |
Hb_086765_010 |
0.2544859588 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 10 |
Hb_017098_010 |
0.259910201 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_004317_060 |
0.2635050823 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_004878_010 |
0.2682389087 |
- |
- |
- |
| 13 |
Hb_002986_060 |
0.2690434018 |
- |
- |
hypothetical protein M569_15879, partial [Genlisea aurea] |
| 14 |
Hb_002477_050 |
0.2708015951 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000139_150 |
0.2768259343 |
- |
- |
transposon protein, putative, Mutator sub-class [Oryza sativa Japonica Group] |
| 16 |
Hb_007456_020 |
0.2784648317 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_041828_010 |
0.2784950616 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 18 |
Hb_008447_030 |
0.2785150451 |
- |
- |
- |
| 19 |
Hb_000575_100 |
0.2796988266 |
- |
- |
PREDICTED: nephrocystin-3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002477_040 |
0.2806023286 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein isoform 2 [Theobroma cacao] |