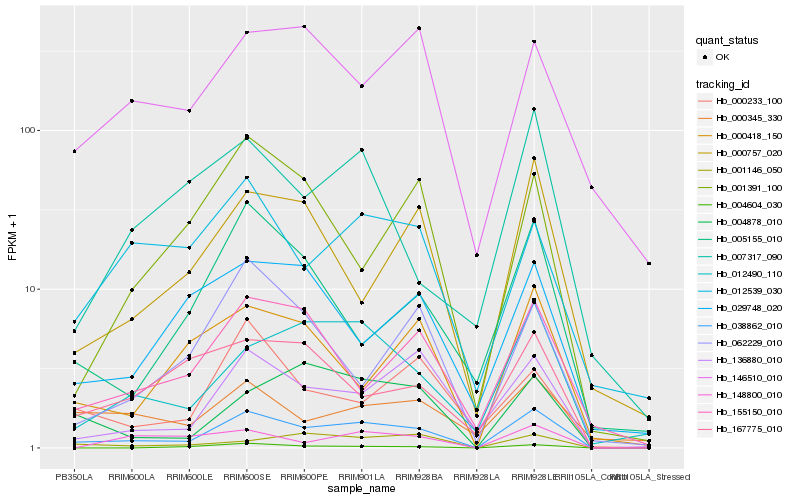
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038862_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_012490_110 |
0.2001678766 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 3 |
Hb_136880_010 |
0.2065648214 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 4 |
Hb_004878_010 |
0.2184813523 |
- |
- |
- |
| 5 |
Hb_000757_020 |
0.2246640761 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_155150_010 |
0.2249435463 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_012539_030 |
0.2252842581 |
- |
- |
PREDICTED: uncharacterized protein LOC105638247 [Jatropha curcas] |
| 8 |
Hb_001146_050 |
0.2252882863 |
- |
- |
- |
| 9 |
Hb_000418_150 |
0.2300096279 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_004604_030 |
0.235000263 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_007317_090 |
0.2377973673 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 12 |
Hb_146510_010 |
0.2385479481 |
- |
- |
cinnamyl alcohol dehydrogenase [Hevea brasiliensis] |
| 13 |
Hb_000345_330 |
0.2388748695 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_167775_010 |
0.2416042589 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_005155_010 |
0.2417222519 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_000233_100 |
0.2428679358 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_029748_020 |
0.2441100799 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001391_100 |
0.2449317302 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 19 |
Hb_062229_010 |
0.2470186106 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 20 |
Hb_148800_010 |
0.2470677106 |
- |
- |
- |