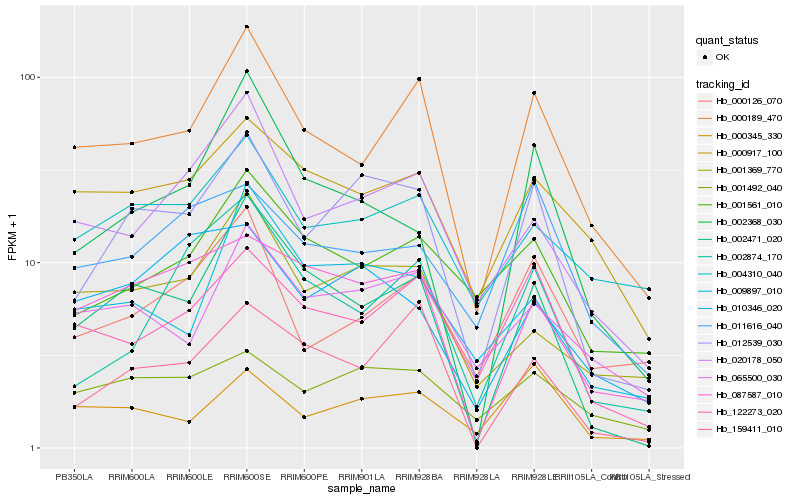
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012539_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638247 [Jatropha curcas] |
| 2 |
Hb_002471_020 |
0.1731636668 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 3 |
Hb_000189_470 |
0.1809384203 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 4 |
Hb_001492_040 |
0.182300243 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_087587_010 |
0.1831811716 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_122273_020 |
0.1842696565 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 7 |
Hb_159411_010 |
0.1901266036 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_020178_050 |
0.1903259083 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 9 |
Hb_000126_070 |
0.1923175761 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 10 |
Hb_000345_330 |
0.1931886775 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_065500_030 |
0.1955568314 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 12 |
Hb_001369_770 |
0.1967734473 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 13 |
Hb_004310_040 |
0.1980625934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009897_010 |
0.1983017624 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_001561_010 |
0.1990083247 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010346_020 |
0.2022626974 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 17 |
Hb_000917_100 |
0.2040398575 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 18 |
Hb_002874_170 |
0.2041525883 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_011616_040 |
0.2043948861 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 20 |
Hb_002368_030 |
0.2047144384 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |