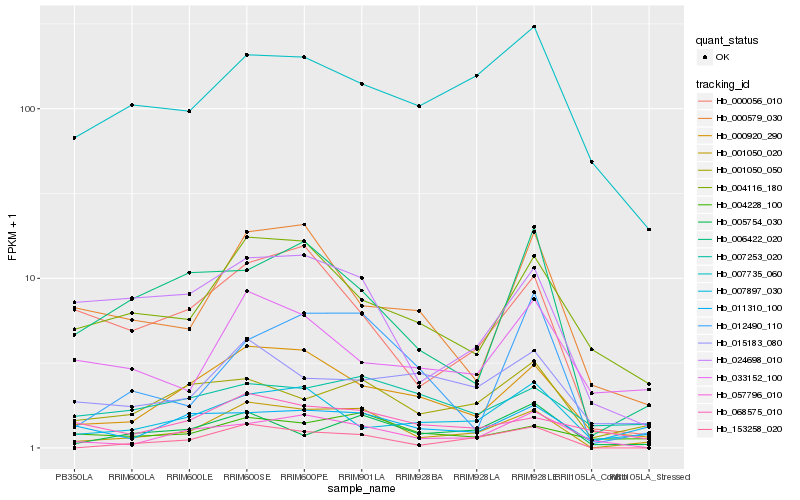
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001050_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000920_290 |
0.1733834997 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_153258_020 |
0.1848026107 |
- |
- |
- |
| 4 |
Hb_001050_050 |
0.2013030204 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g57250, mitochondrial [Jatropha curcas] |
| 5 |
Hb_004116_180 |
0.2093656693 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 6 |
Hb_005754_030 |
0.2133799965 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 7 |
Hb_024698_010 |
0.2135849435 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46 [Jatropha curcas] |
| 8 |
Hb_004228_100 |
0.2155015712 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 9 |
Hb_012490_110 |
0.2167736345 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 10 |
Hb_000056_010 |
0.2190732746 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 11 |
Hb_015183_080 |
0.2240428398 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_007897_030 |
0.2243239166 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 13 |
Hb_006422_020 |
0.2265730202 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 14 |
Hb_007735_060 |
0.2270820775 |
- |
- |
PPase [Hevea brasiliensis] |
| 15 |
Hb_057796_010 |
0.2275560328 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 16 |
Hb_011310_100 |
0.2308296749 |
- |
- |
hypothetical protein CICLE_v10008199mg [Citrus clementina] |
| 17 |
Hb_000579_030 |
0.2317066222 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 18 |
Hb_007253_020 |
0.2321145253 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 19 |
Hb_068575_010 |
0.2327057436 |
- |
- |
PREDICTED: protein GAMETE EXPRESSED 2 [Jatropha curcas] |
| 20 |
Hb_033152_100 |
0.2345951267 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |