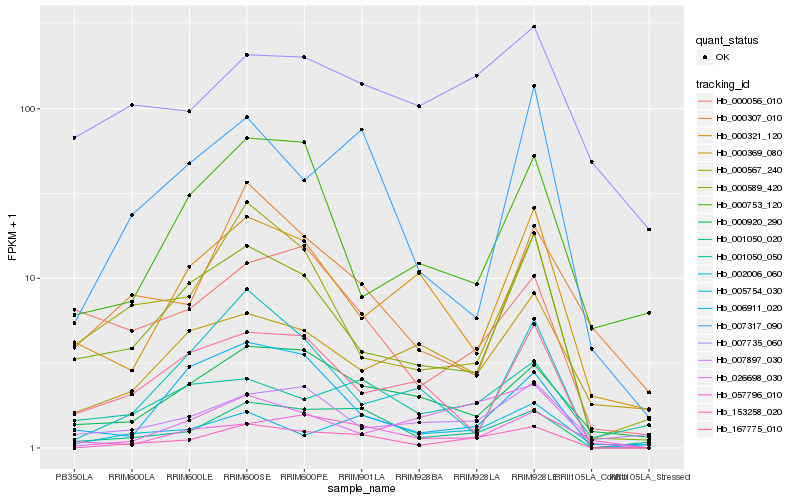
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153258_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_001050_020 |
0.1848026107 |
- |
- |
- |
| 3 |
Hb_005754_030 |
0.2407030636 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 4 |
Hb_000920_290 |
0.2428307566 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_026698_030 |
0.2437384358 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 6 |
Hb_007317_090 |
0.245007467 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 7 |
Hb_000589_420 |
0.2487938154 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 8 |
Hb_057796_010 |
0.2508349234 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 9 |
Hb_006911_020 |
0.26436476 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000567_240 |
0.2659895898 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 11 |
Hb_002006_060 |
0.2666498887 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 12 |
Hb_007735_060 |
0.2679540555 |
- |
- |
PPase [Hevea brasiliensis] |
| 13 |
Hb_007897_030 |
0.2682321748 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 14 |
Hb_001050_050 |
0.2737837018 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g57250, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000056_010 |
0.2761296268 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 16 |
Hb_000307_010 |
0.2772897104 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 17 |
Hb_167775_010 |
0.2778404847 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_000369_080 |
0.2812568028 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 19 |
Hb_000321_120 |
0.2821332873 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 20 |
Hb_000753_120 |
0.2841720598 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |