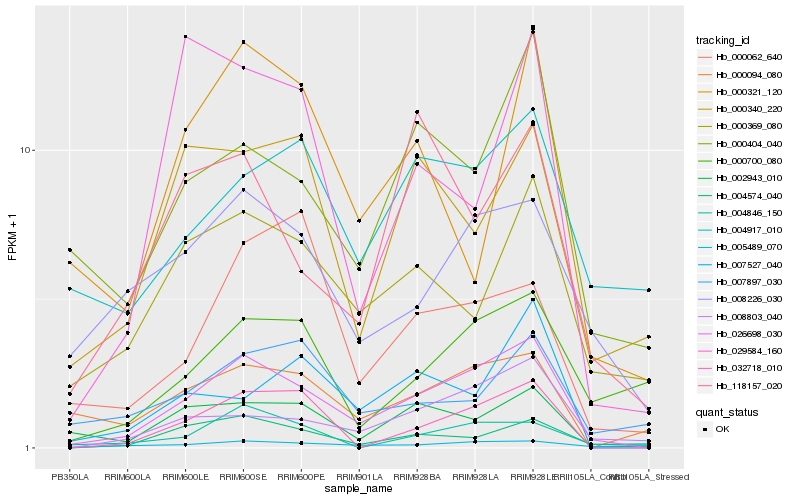
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026698_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 2 |
Hb_008803_040 |
0.1866868829 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |
| 3 |
Hb_002943_010 |
0.2038323212 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 4 |
Hb_008226_030 |
0.2040636327 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 5 |
Hb_000404_040 |
0.2054722253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_032718_010 |
0.2073323014 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 7 |
Hb_004574_040 |
0.2092032899 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 8 |
Hb_000369_080 |
0.2120380922 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 9 |
Hb_000094_080 |
0.212828314 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 10 |
Hb_000700_080 |
0.2209596447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007527_040 |
0.2244522426 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 12 |
Hb_005489_070 |
0.2251746179 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 13 |
Hb_004846_150 |
0.2256811811 |
- |
- |
hypothetical protein JCGZ_26968 [Jatropha curcas] |
| 14 |
Hb_118157_020 |
0.2262038975 |
- |
- |
hypothetical protein MTR_3g456110 [Medicago truncatula] |
| 15 |
Hb_000340_220 |
0.2264029479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000062_640 |
0.2348745114 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 17 |
Hb_029584_160 |
0.2353469265 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 18 |
Hb_004917_010 |
0.2355411863 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 19 |
Hb_000321_120 |
0.235739725 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 20 |
Hb_007897_030 |
0.2361314029 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |