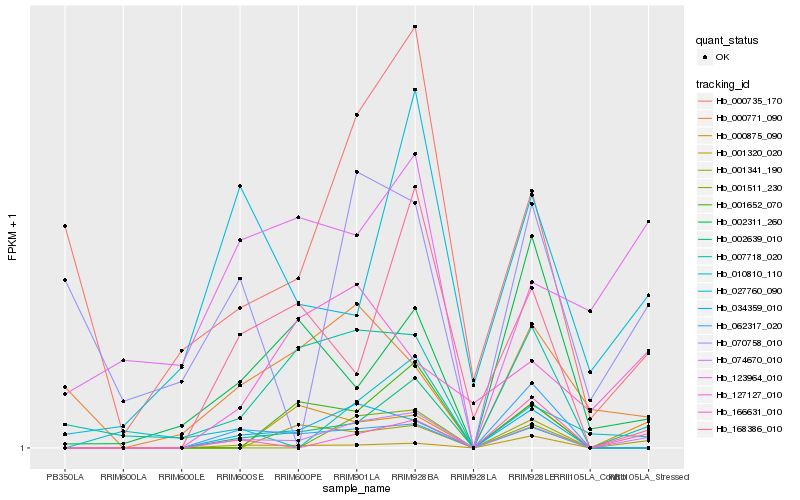
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074670_010 |
0.0 |
- |
- |
hypothetical protein VITISV_015625 [Vitis vinifera] |
| 2 |
Hb_002639_010 |
0.2726343667 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 3 |
Hb_034359_010 |
0.27902886 |
- |
- |
- |
| 4 |
Hb_007718_020 |
0.2836356914 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 5 |
Hb_166631_010 |
0.3068942 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 6 |
Hb_168386_010 |
0.3100093429 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_001341_190 |
0.3118418835 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000735_170 |
0.3159651483 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 9 |
Hb_027760_090 |
0.3291388011 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000875_090 |
0.3353204288 |
- |
- |
hypothetical protein VITISV_005326 [Vitis vinifera] |
| 11 |
Hb_010810_110 |
0.3437493094 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_001652_070 |
0.3504988743 |
- |
- |
PREDICTED: uncharacterized protein LOC103439285 [Malus domestica] |
| 13 |
Hb_001511_230 |
0.3556230969 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 14 |
Hb_127127_010 |
0.3575778747 |
- |
- |
- |
| 15 |
Hb_002311_260 |
0.3593114039 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 16 |
Hb_001320_020 |
0.3611626078 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 17 |
Hb_123964_010 |
0.3655351597 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 18 |
Hb_062317_020 |
0.367951412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000771_090 |
0.3690774644 |
- |
- |
hypothetical protein B456_012G003500 [Gossypium raimondii] |
| 20 |
Hb_070758_010 |
0.3706852622 |
- |
- |
- |