| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
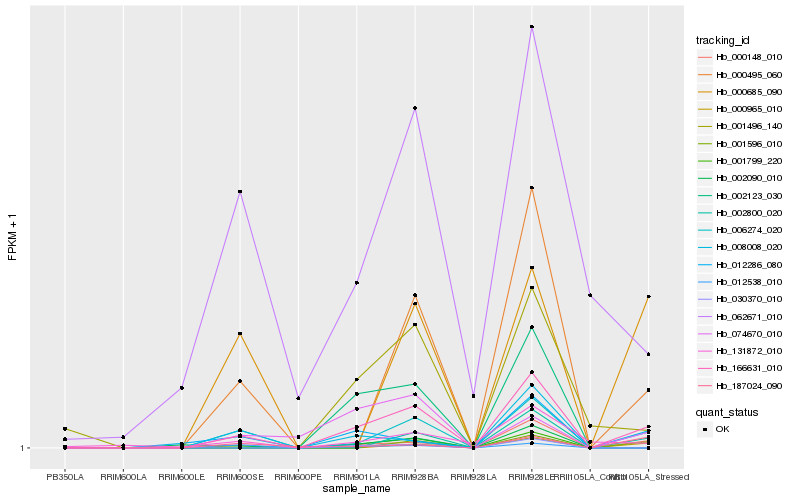
Hb_166631_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 2 |
Hb_008008_020 |
0.2439325644 |
- |
- |
- |
| 3 |
Hb_000148_010 |
0.2519580299 |
- |
- |
- |
| 4 |
Hb_000495_060 |
0.2561161388 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 5 |
Hb_012286_080 |
0.2656909888 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 6 |
Hb_000965_010 |
0.2778490292 |
- |
- |
hypothetical protein VITISV_016917 [Vitis vinifera] |
| 7 |
Hb_006274_020 |
0.3018815746 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 8 |
Hb_074670_010 |
0.3068942 |
- |
- |
hypothetical protein VITISV_015625 [Vitis vinifera] |
| 9 |
Hb_001496_140 |
0.3117790951 |
- |
- |
- |
| 10 |
Hb_001799_220 |
0.3151343917 |
transcription factor |
TF Family: AP2 |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 11 |
Hb_002800_020 |
0.3162895958 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_002090_010 |
0.3177468627 |
- |
- |
gag-pol fusion protein [Rhizophagus irregularis DAOM 197198w] |
| 13 |
Hb_030370_010 |
0.3201410784 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_131872_010 |
0.3214259952 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_002123_030 |
0.3235895117 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 16 |
Hb_000685_090 |
0.3239985809 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 17 |
Hb_187024_090 |
0.3245576908 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_001596_010 |
0.3252592696 |
- |
- |
hypothetical protein POPTR_0001s181102g, partial [Populus trichocarpa] |
| 19 |
Hb_012538_010 |
0.3266348825 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_062671_010 |
0.3271997055 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |