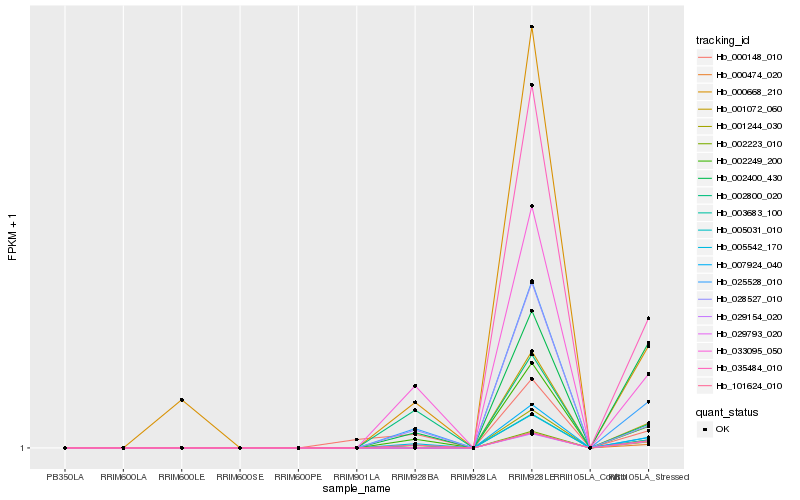
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_170 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_025528_010 |
0.0365464695 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 3 |
Hb_002249_200 |
0.0473030371 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |
| 4 |
Hb_005031_010 |
0.0826387613 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 5 |
Hb_101624_010 |
0.0826992687 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 6 |
Hb_033095_050 |
0.0887934757 |
- |
- |
hypothetical protein VITISV_038692 [Vitis vinifera] |
| 7 |
Hb_001244_030 |
0.1237684242 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_028527_010 |
0.1408956444 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 9 |
Hb_002400_430 |
0.1747479673 |
- |
- |
- |
| 10 |
Hb_002800_020 |
0.1832309079 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_000668_210 |
0.1996447234 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |
| 12 |
Hb_035484_010 |
0.210539085 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 13 |
Hb_000148_010 |
0.2115073863 |
- |
- |
- |
| 14 |
Hb_001072_060 |
0.2145579738 |
- |
- |
PREDICTED: uncharacterized protein LOC105645525 [Jatropha curcas] |
| 15 |
Hb_007924_040 |
0.2156061449 |
- |
- |
PREDICTED: pollen-specific leucine-rich repeat extensin-like protein 3 [Populus euphratica] |
| 16 |
Hb_003683_100 |
0.2157927409 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_000474_020 |
0.2161562394 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 18 |
Hb_029154_020 |
0.2235903181 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_029793_020 |
0.2236308492 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_002223_010 |
0.2237669793 |
- |
- |
hypothetical protein VITISV_009489 [Vitis vinifera] |