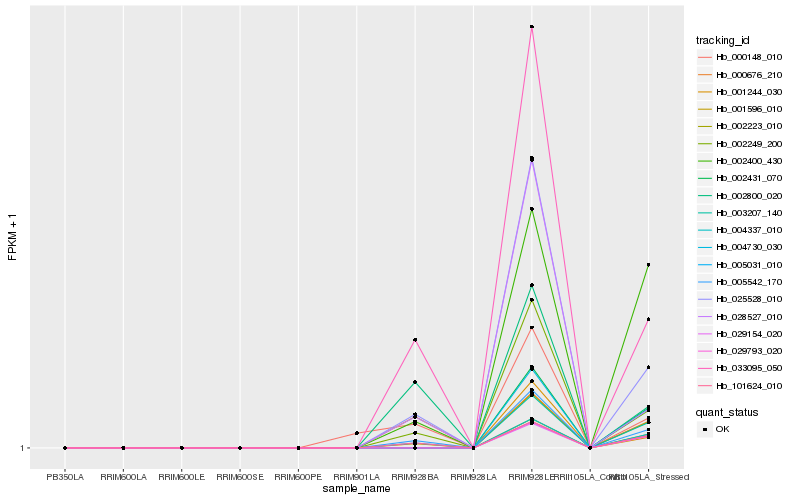
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101624_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_005031_010 |
6.15271e-05 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 3 |
Hb_005542_170 |
0.0826992687 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_033095_050 |
0.1070407277 |
- |
- |
hypothetical protein VITISV_038692 [Vitis vinifera] |
| 5 |
Hb_025528_010 |
0.1189972122 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 6 |
Hb_002400_430 |
0.1281162054 |
- |
- |
- |
| 7 |
Hb_002249_200 |
0.1297612277 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |
| 8 |
Hb_002800_020 |
0.1792857585 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_001596_010 |
0.1803513124 |
- |
- |
hypothetical protein POPTR_0001s181102g, partial [Populus trichocarpa] |
| 10 |
Hb_001244_030 |
0.2054199064 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_028527_010 |
0.2170731466 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 12 |
Hb_000148_010 |
0.228701675 |
- |
- |
- |
| 13 |
Hb_029793_020 |
0.2418709841 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_029154_020 |
0.2418710629 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_002223_010 |
0.2418711493 |
- |
- |
hypothetical protein VITISV_009489 [Vitis vinifera] |
| 16 |
Hb_004337_010 |
0.2418711675 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_003207_140 |
0.2418909875 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 18 |
Hb_000676_210 |
0.2418948116 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 19 |
Hb_004730_030 |
0.2419540386 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 20 |
Hb_002431_070 |
0.2419609502 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |