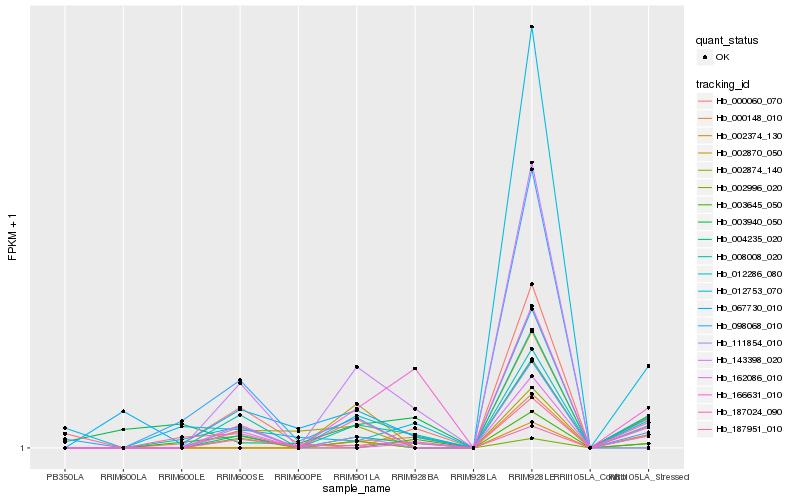
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012286_080 |
0.0 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 2 |
Hb_008008_020 |
0.1669433414 |
- |
- |
- |
| 3 |
Hb_111854_010 |
0.25704655 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 4 |
Hb_166631_010 |
0.2656909888 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 5 |
Hb_187951_010 |
0.3057869048 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_000148_010 |
0.3101889001 |
- |
- |
- |
| 7 |
Hb_143398_020 |
0.3158976287 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 8 |
Hb_098068_010 |
0.3184167054 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 9 |
Hb_000060_070 |
0.3192165951 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 10 |
Hb_067730_010 |
0.320085399 |
- |
- |
hypothetical protein OsJ_21419 [Oryza sativa Japonica Group] |
| 11 |
Hb_187024_090 |
0.3255663849 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_002870_050 |
0.3285771904 |
- |
- |
- |
| 13 |
Hb_002874_140 |
0.3342094884 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |
| 14 |
Hb_002996_020 |
0.335069291 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_003645_050 |
0.3350765987 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_162086_010 |
0.3378348937 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 17 |
Hb_002374_130 |
0.3402391116 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 18 |
Hb_003940_050 |
0.3427505867 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 19 |
Hb_004235_020 |
0.3462511651 |
- |
- |
PREDICTED: uncharacterized protein LOC103322555 [Prunus mume] |
| 20 |
Hb_012753_070 |
0.3490391641 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |