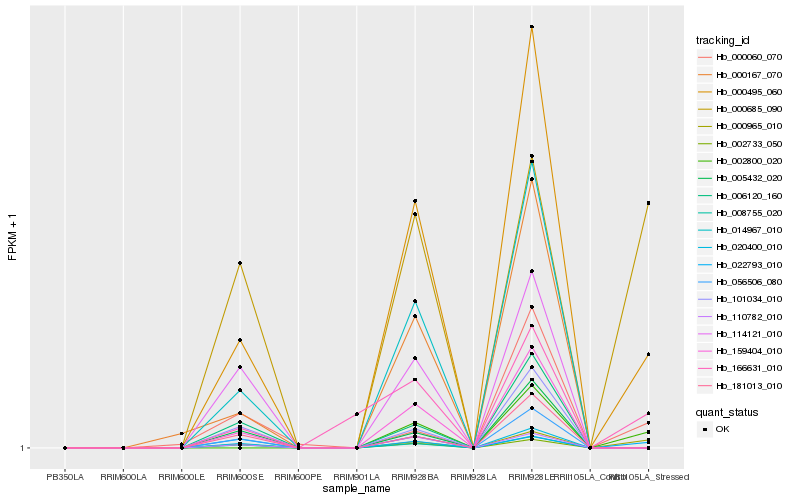
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000495_060 |
0.0 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 2 |
Hb_000965_010 |
0.179751126 |
- |
- |
hypothetical protein VITISV_016917 [Vitis vinifera] |
| 3 |
Hb_014967_010 |
0.2093137479 |
- |
- |
- |
| 4 |
Hb_159404_010 |
0.211417747 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 5 |
Hb_020400_010 |
0.2116543132 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 6 |
Hb_008755_020 |
0.2219633711 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_110782_010 |
0.225079509 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 8 |
Hb_056506_080 |
0.2314208429 |
- |
- |
PREDICTED: uncharacterized protein LOC104888944 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_022793_010 |
0.2389093237 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 10 |
Hb_002800_020 |
0.2396110519 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_002733_050 |
0.2437443926 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_114121_010 |
0.2438719692 |
- |
- |
- |
| 13 |
Hb_000167_070 |
0.2448597601 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 14 |
Hb_006120_160 |
0.2531315397 |
- |
- |
- |
| 15 |
Hb_000685_090 |
0.2535671451 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 16 |
Hb_101034_010 |
0.2549659891 |
- |
- |
- |
| 17 |
Hb_000060_070 |
0.2561139401 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 18 |
Hb_166631_010 |
0.2561161388 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 19 |
Hb_005432_020 |
0.2566445962 |
- |
- |
- |
| 20 |
Hb_181013_010 |
0.258536601 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |