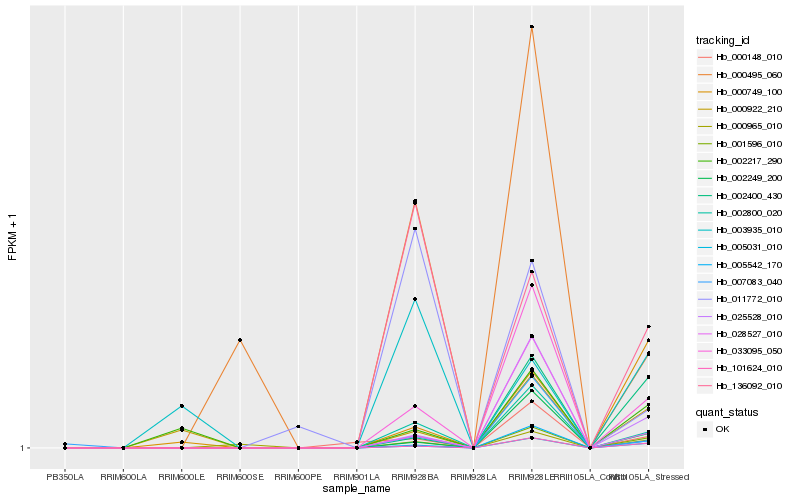
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001596_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s181102g, partial [Populus trichocarpa] |
| 2 |
Hb_002800_020 |
0.113491352 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_033095_050 |
0.1628086228 |
- |
- |
hypothetical protein VITISV_038692 [Vitis vinifera] |
| 4 |
Hb_101624_010 |
0.1803513124 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_005031_010 |
0.1803869496 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 6 |
Hb_136092_010 |
0.1815275628 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 7 |
Hb_005542_170 |
0.2283769393 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_000965_010 |
0.2452291097 |
- |
- |
hypothetical protein VITISV_016917 [Vitis vinifera] |
| 9 |
Hb_025528_010 |
0.2527921894 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 10 |
Hb_002249_200 |
0.2620349777 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |
| 11 |
Hb_000148_010 |
0.2712128326 |
- |
- |
- |
| 12 |
Hb_011772_010 |
0.2768901169 |
- |
- |
- |
| 13 |
Hb_000495_060 |
0.2775936924 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 14 |
Hb_002400_430 |
0.2777489936 |
- |
- |
- |
| 15 |
Hb_000922_210 |
0.2956265075 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002217_290 |
0.2956678321 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 17 |
Hb_028527_010 |
0.3046675617 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 18 |
Hb_000749_100 |
0.3090747916 |
- |
- |
- |
| 19 |
Hb_003935_010 |
0.3103219873 |
- |
- |
- |
| 20 |
Hb_007083_040 |
0.3164172375 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |