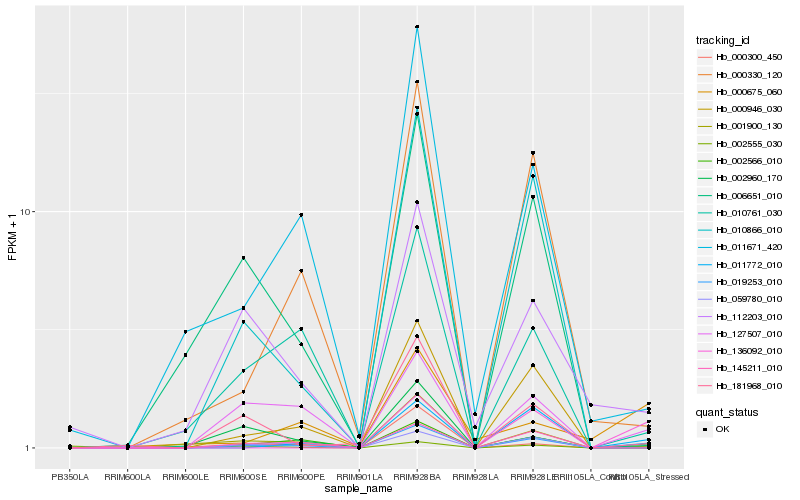
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019253_010 |
0.0 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 2 |
Hb_127507_010 |
0.2246627788 |
- |
- |
PREDICTED: uncharacterized protein LOC104814513 [Tarenaya hassleriana] |
| 3 |
Hb_011772_010 |
0.2253206444 |
- |
- |
- |
| 4 |
Hb_001900_130 |
0.2407863003 |
- |
- |
PREDICTED: uncharacterized protein LOC105632667 [Jatropha curcas] |
| 5 |
Hb_000946_030 |
0.2482084089 |
- |
- |
hypothetical protein JCGZ_10707 [Jatropha curcas] |
| 6 |
Hb_000330_120 |
0.2555246451 |
- |
- |
PREDICTED: bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase-like [Nelumbo nucifera] |
| 7 |
Hb_010866_010 |
0.2563130295 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 8 |
Hb_000300_450 |
0.2590846748 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_010761_030 |
0.2593817052 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_181968_010 |
0.2632599779 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 11 |
Hb_136092_010 |
0.2653208377 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 12 |
Hb_011671_420 |
0.2674243841 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 13 |
Hb_145211_010 |
0.2687892463 |
- |
- |
hypothetical protein M569_05648, partial [Genlisea aurea] |
| 14 |
Hb_000675_060 |
0.275041069 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_002566_010 |
0.2762988424 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002555_030 |
0.2769320629 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 17 |
Hb_112203_010 |
0.2772798469 |
- |
- |
- |
| 18 |
Hb_002960_170 |
0.2804519613 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 19 |
Hb_059780_010 |
0.2834992991 |
- |
- |
PREDICTED: uncharacterized protein LOC104885482, partial [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_006651_010 |
0.2848795715 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |