| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
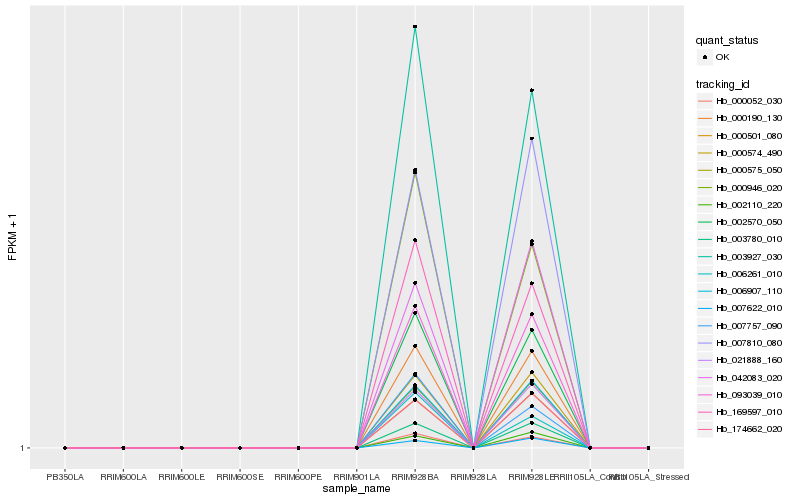
Hb_002570_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104897677 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_093039_010 |
0.0183667618 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 3 |
Hb_000190_130 |
0.0220310963 |
- |
- |
cysteine protease inhibitor [Populus tremula] |
| 4 |
Hb_000052_030 |
0.0232965575 |
- |
- |
- |
| 5 |
Hb_003927_030 |
0.0246357344 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_169597_010 |
0.0298426326 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_003780_010 |
0.038578905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000574_490 |
0.0442654008 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 11 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006907_110 |
0.0520893398 |
- |
- |
- |
| 10 |
Hb_000575_050 |
0.0529884522 |
- |
- |
hypothetical protein CISIN_1g0468601mg, partial [Citrus sinensis] |
| 11 |
Hb_000946_020 |
0.0548865149 |
- |
- |
hypothetical protein PRUPE_ppa023262mg [Prunus persica] |
| 12 |
Hb_021888_160 |
0.0553391193 |
- |
- |
- |
| 13 |
Hb_000501_080 |
0.0630741638 |
- |
- |
hypothetical protein OsJ_22457 [Oryza sativa Japonica Group] |
| 14 |
Hb_174662_020 |
0.0634049319 |
- |
- |
PREDICTED: uncharacterized protein LOC104888075 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_007810_080 |
0.0683881916 |
- |
- |
PREDICTED: uncharacterized protein LOC103491654 [Cucumis melo] |
| 16 |
Hb_006261_010 |
0.0897400094 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 17 |
Hb_002110_220 |
0.0920049852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007622_010 |
0.0957774689 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 19 |
Hb_007757_090 |
0.0961325209 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |
| 20 |
Hb_042083_020 |
0.0966480056 |
- |
- |
- |