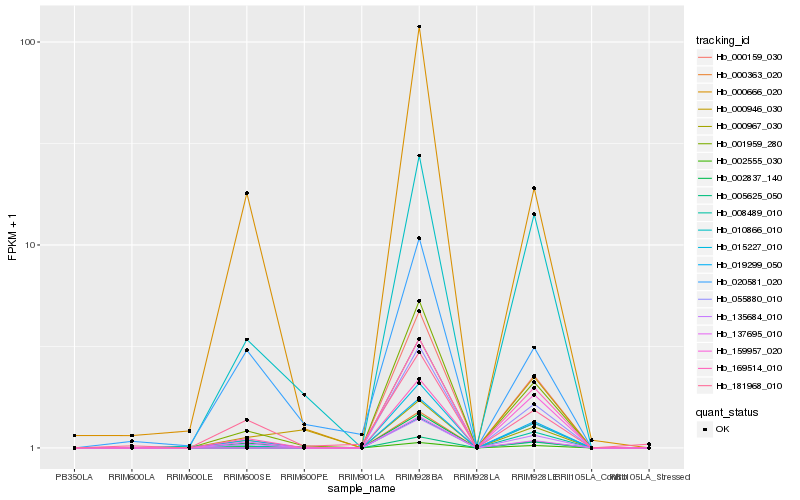
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135684_010 |
0.0 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 2 |
Hb_015227_010 |
0.0558496631 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_000363_020 |
0.0998242555 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_001959_280 |
0.111445273 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 5 |
Hb_010866_010 |
0.1139995545 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 6 |
Hb_137695_010 |
0.1280752923 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Malus domestica] |
| 7 |
Hb_169514_010 |
0.1525205606 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 8 |
Hb_008489_010 |
0.1537381824 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 9 |
Hb_002837_140 |
0.159140311 |
- |
- |
hypothetical protein JCGZ_05022 [Jatropha curcas] |
| 10 |
Hb_000666_020 |
0.167125226 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 11 |
Hb_181968_010 |
0.1782066428 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 12 |
Hb_000946_030 |
0.1931759365 |
- |
- |
hypothetical protein JCGZ_10707 [Jatropha curcas] |
| 13 |
Hb_002555_030 |
0.194139275 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 14 |
Hb_020581_020 |
0.1961190165 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 15 |
Hb_000967_030 |
0.1973611016 |
- |
- |
- |
| 16 |
Hb_000159_030 |
0.1988597429 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 17 |
Hb_159957_020 |
0.1990840723 |
- |
- |
- |
| 18 |
Hb_019299_050 |
0.1995376367 |
- |
- |
- |
| 19 |
Hb_055880_010 |
0.203526382 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_005625_050 |
0.2039108391 |
- |
- |
hypothetical protein PRUPE_ppa024499mg, partial [Prunus persica] |