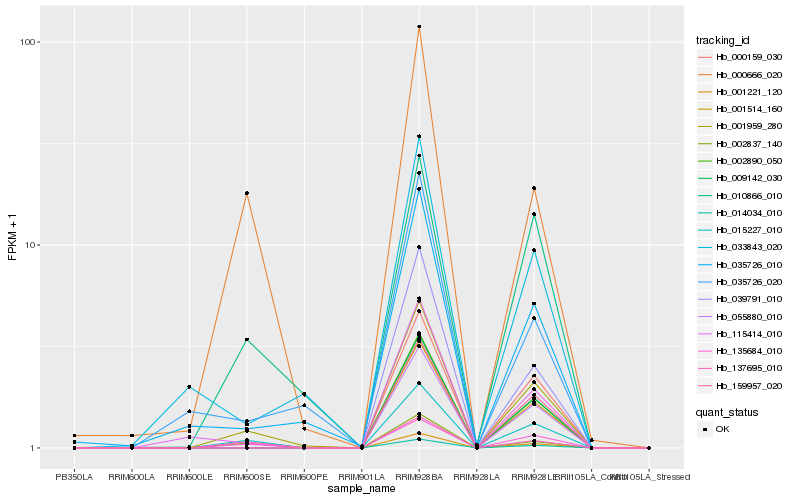
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 2 |
Hb_015227_010 |
0.0679337503 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_135684_010 |
0.111445273 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 4 |
Hb_002837_140 |
0.1194759462 |
- |
- |
hypothetical protein JCGZ_05022 [Jatropha curcas] |
| 5 |
Hb_035726_010 |
0.1308190842 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 6 |
Hb_010866_010 |
0.1408688252 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 7 |
Hb_115414_010 |
0.1447395862 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 8 |
Hb_001221_120 |
0.1461716954 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_137695_010 |
0.1462671953 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Malus domestica] |
| 10 |
Hb_002890_050 |
0.1463906548 |
- |
- |
- |
| 11 |
Hb_014034_010 |
0.1464880577 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_001514_160 |
0.1470471907 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 13 |
Hb_009142_030 |
0.1473207605 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_055880_010 |
0.1477920688 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000666_020 |
0.151766134 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 16 |
Hb_159957_020 |
0.1533936891 |
- |
- |
- |
| 17 |
Hb_000159_030 |
0.1539303496 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 18 |
Hb_033843_020 |
0.156686593 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 19 |
Hb_039791_010 |
0.1575061797 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_035726_020 |
0.160224576 |
- |
- |
Insulinase (Peptidase family M16) family protein [Theobroma cacao] |