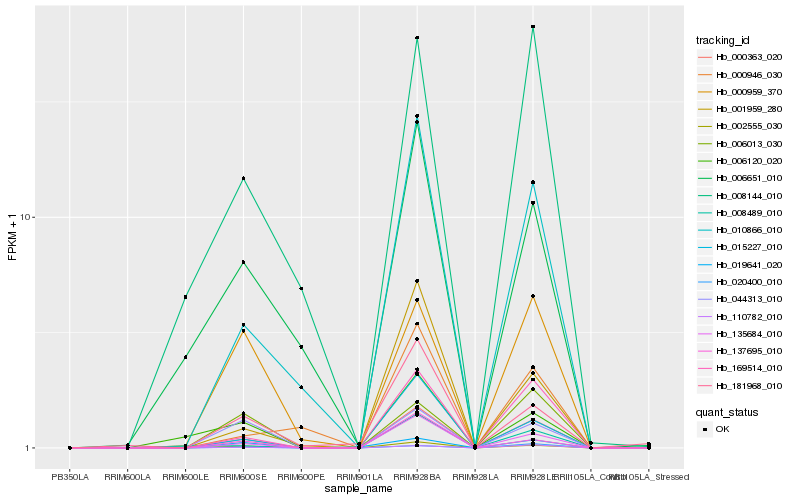
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_020 |
0.0 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_169514_010 |
0.0963430191 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 3 |
Hb_135684_010 |
0.0998242555 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 4 |
Hb_010866_010 |
0.1302397629 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 5 |
Hb_110782_010 |
0.1469867261 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 6 |
Hb_015227_010 |
0.1551821389 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_008489_010 |
0.1765320938 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_002555_030 |
0.1859599255 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 9 |
Hb_008144_010 |
0.1952601523 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 10 |
Hb_020400_010 |
0.2016447874 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 11 |
Hb_001959_280 |
0.2024932115 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 12 |
Hb_137695_010 |
0.2025422998 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Malus domestica] |
| 13 |
Hb_000959_370 |
0.2034440117 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 14 |
Hb_019641_020 |
0.2087399392 |
- |
- |
hypothetical protein AALP_AAs49670U000100, partial [Arabis alpina] |
| 15 |
Hb_000946_030 |
0.2096256788 |
- |
- |
hypothetical protein JCGZ_10707 [Jatropha curcas] |
| 16 |
Hb_006013_030 |
0.2098525268 |
- |
- |
- |
| 17 |
Hb_044313_010 |
0.2134108115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006120_020 |
0.2140978113 |
- |
- |
- |
| 19 |
Hb_006651_010 |
0.2142106184 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 20 |
Hb_181968_010 |
0.2166392011 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |