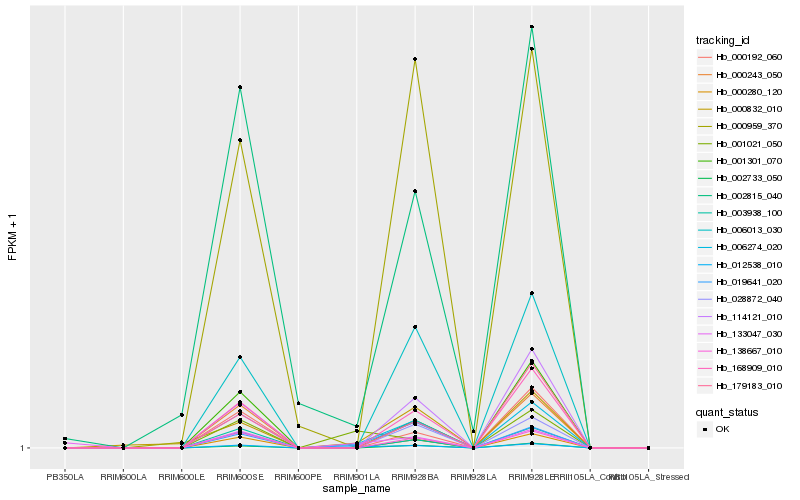
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012538_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_006274_020 |
0.1544984844 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 3 |
Hb_001021_050 |
0.1802801006 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_019641_020 |
0.1816201676 |
- |
- |
hypothetical protein AALP_AAs49670U000100, partial [Arabis alpina] |
| 5 |
Hb_000832_010 |
0.2124909244 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 6 |
Hb_133047_030 |
0.2245800115 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000280_120 |
0.2266365453 |
- |
- |
PREDICTED: uncharacterized protein LOC105643934 [Jatropha curcas] |
| 8 |
Hb_168909_010 |
0.2290824949 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 9 |
Hb_000243_050 |
0.2314236296 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 10 |
Hb_138667_010 |
0.2328794124 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 11 |
Hb_002733_050 |
0.2363492501 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_114121_010 |
0.2374309277 |
- |
- |
- |
| 13 |
Hb_006013_030 |
0.2376284497 |
- |
- |
- |
| 14 |
Hb_002815_040 |
0.2406418789 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 15 |
Hb_028872_040 |
0.2447965064 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_001301_070 |
0.2488059978 |
- |
- |
- |
| 17 |
Hb_003938_100 |
0.2508028071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000959_370 |
0.2589225811 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 19 |
Hb_179183_010 |
0.2592295285 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 20 |
Hb_000192_060 |
0.2594314417 |
- |
- |
- |