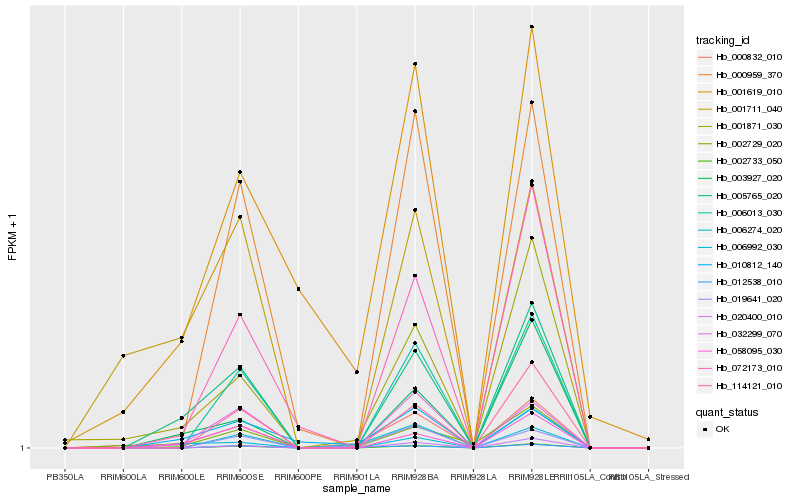
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000832_010 |
0.0 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 2 |
Hb_001871_030 |
0.1378100339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006274_020 |
0.1633063805 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_002729_020 |
0.1756697997 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 5 |
Hb_005765_020 |
0.1854223853 |
- |
- |
- |
| 6 |
Hb_019641_020 |
0.1905285808 |
- |
- |
hypothetical protein AALP_AAs49670U000100, partial [Arabis alpina] |
| 7 |
Hb_072173_010 |
0.2077367807 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 8 |
Hb_006013_030 |
0.2079177506 |
- |
- |
- |
| 9 |
Hb_032299_070 |
0.2091446243 |
- |
- |
- |
| 10 |
Hb_002733_050 |
0.2100597252 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_012538_010 |
0.2124909244 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_010812_140 |
0.2133144706 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_001619_010 |
0.21413215 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 14 |
Hb_000959_370 |
0.216511538 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 15 |
Hb_058095_030 |
0.2178769707 |
- |
- |
- |
| 16 |
Hb_006992_030 |
0.2181733085 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_114121_010 |
0.2187224594 |
- |
- |
- |
| 18 |
Hb_003927_020 |
0.2210636359 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 19 |
Hb_001711_040 |
0.2260555595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_020400_010 |
0.2265086372 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |