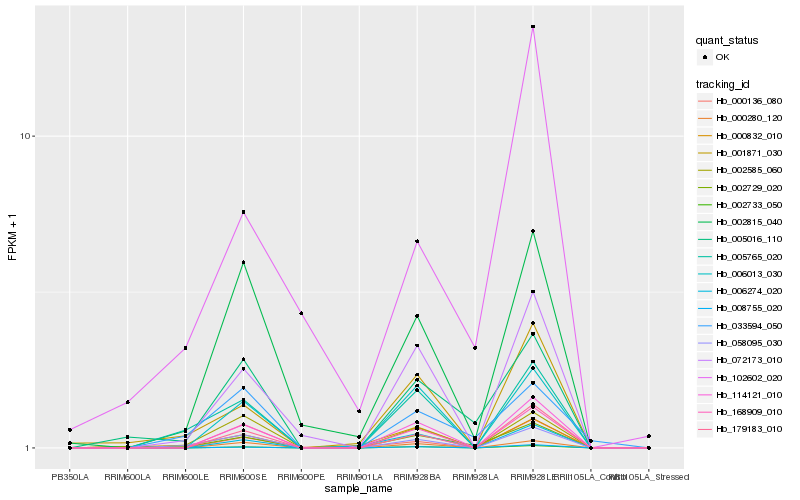
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002729_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 2 |
Hb_005016_110 |
0.122894437 |
- |
- |
- |
| 3 |
Hb_000832_010 |
0.1756697997 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 4 |
Hb_001871_030 |
0.1926224173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000136_080 |
0.2174905468 |
- |
- |
hypothetical protein AALP_AA8G499800 [Arabis alpina] |
| 6 |
Hb_002815_040 |
0.2201227974 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 7 |
Hb_114121_010 |
0.2252936927 |
- |
- |
- |
| 8 |
Hb_002733_050 |
0.2265193158 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_005765_020 |
0.2290872616 |
- |
- |
- |
| 10 |
Hb_058095_030 |
0.2295085517 |
- |
- |
- |
| 11 |
Hb_072173_010 |
0.2300540663 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 12 |
Hb_006274_020 |
0.2309185294 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 13 |
Hb_168909_010 |
0.2316092717 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 14 |
Hb_006013_030 |
0.236926829 |
- |
- |
- |
| 15 |
Hb_008755_020 |
0.238287731 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_179183_010 |
0.241820782 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 17 |
Hb_002585_060 |
0.242755285 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_033594_050 |
0.2432243102 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 19 |
Hb_102602_020 |
0.2451619035 |
- |
- |
hypothetical protein B456_001G166300 [Gossypium raimondii] |
| 20 |
Hb_000280_120 |
0.2468038212 |
- |
- |
PREDICTED: uncharacterized protein LOC105643934 [Jatropha curcas] |